Gene
KWMTBOMO15743
Pre Gene Modal
BGIBMGA009444
Annotation
PREDICTED:_uncharacterized_protein_LOC105842932_[Bombyx_mori]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 1.163 PlasmaMembrane Reliability : 1.069
Sequence
CDS
ATGACAACCAGCAAAATCCAAGGACAAATATATCATTTGCATGGTTCAATGGTGCCAACACCAAATGAACCGCATCAATTTCTGCAAATATATTTCATTTCGTCGATGGTGGATCAGCTGAATGTGCGGTGCAATATACAGGGAACACAACAGTTAAAGAGACGAATTATTGAACAGTTGCAAGCATTTTTTCACGCTAATAATGCTGTGGTTAATATGTTCAAAACAGCATTGGAACGAATGCCATCGGATACGCACAAATTTGTCATAAGAGCGGATTGTACCCCAACAGGGGAACATGTGCGAAGATTCAATGCACCCACCGTTAATGATGTTGCTGCTATTATTGTTGGCGATCCAACTAAATCGCGAGACATTGTCGTTCAGCGAAGAAGCAATATCATGCATCGTGTAAACGAGACACATCGTTTGTACGATGCGTTACAATATCCAATCATTTATTGGCAAGGACAAGACGGATACGACATCACGTTGAAGATGGTCGATCCAATTACAGGCGTATCAACGAATAAAAATCTAAGCGCAATGAATTACTATGCGTATCGTATGATGATTCGTACACATGAGGAGAATGTCATTCTGAAGTGCCGTCGGCTATTCCAGCAATTCGCTGTCGACATGTATGTCAAAGTCGAGACCGAACGTTTAGCGTTCATCCGATTCAATCAGGCAAAGCTACGATCTGAGGACTATATACACTTGCGTGATGCTATTCATTCAGATGTTGTACATTTAGCAGTGCATTTGGAAAATGGCCAAAGAGTTTACTTCACTGAGGCTAATGCGGCACAACGAGCTGAAAGACCACCATCGACAACATTGACTAGCTTCTTTGCAATGTGTGAAGCAGATCCATTCGCAGCGACGCTGATGTACGTTGAAATGCCCAAGTATTACACTTGGAATCAATCAACAAAGAAATTCCAACGTCGCAAACAAGGAACCCCAGTTCCAGATTGGCCACAGGTGTTTTCCACTGATGCACTAGGTCGCATGTATACTGTTCATCCTAGAAATGACGAATGTTTTTATTTGCGACTGCTGCTGGTAAATGTACGTGGACCAAAATCATTTGCGCATTTGAAAACTGTGAATGGCCACCAATGCCAAACATATCGAGAAGCATGTCAACTATTGGGTTTGCTGGAGAACGATTCTCATTGGGATTTAACACTTGCGGATTCAGTTGTTTCATCAAATGCGTACCAAATACGAACGCTGTTCGCAATTATCATCACCACATGTTTTCCTTCACAACCAATTCAGTTATGGAACAAATTCAAAGACGACATATGTGAAGATATCTTGCATCGCTTGCGCATTCAAACGAATAATCCTGACATCCAAATAACCGATGAAATCTACAATGAAGGATTGATTCTGATTGAGGATCAATGCTTGACTATTGCAAACAAGTTACTGATTGAAGTAGGAATGATTGCGCCAAATCGATCGATGCACGATGCATTCAACCAAGAATTAAATCGAGAGCTGCAATACAATGTTGATACATTGCAGGAATTCGTTCGAAATAATGTGCCGTTACTGAATGAACAGCAAAAACAAGTATACGAAACATTAATGCAAGCGGTGGACAATAATACTGGTGGTCTATTCTTCCTGGACGCACCTGGAGGAACAGGGAAAACATTTGTCATTTCATTGATTTTGGCCACTATTCGATCAAGATGTGACATAGCTTTGGCGTTAGCATCATCTGGAATTGCGGCGACTCTTCTAGACGGCGGTCAGCACTTCAATGAAACGGATAATTTGCGGACACGTATAACGCTTCGATTCAGTATTTACTTTGGATTCACTTTCATTTACTTAGAGAAGTTCAACTAA
Protein
MTTSKIQGQIYHLHGSMVPTPNEPHQFLQIYFISSMVDQLNVRCNIQGTQQLKRRIIEQLQAFFHANNAVVNMFKTALERMPSDTHKFVIRADCTPTGEHVRRFNAPTVNDVAAIIVGDPTKSRDIVVQRRSNIMHRVNETHRLYDALQYPIIYWQGQDGYDITLKMVDPITGVSTNKNLSAMNYYAYRMMIRTHEENVILKCRRLFQQFAVDMYVKVETERLAFIRFNQAKLRSEDYIHLRDAIHSDVVHLAVHLENGQRVYFTEANAAQRAERPPSTTLTSFFAMCEADPFAATLMYVEMPKYYTWNQSTKKFQRRKQGTPVPDWPQVFSTDALGRMYTVHPRNDECFYLRLLLVNVRGPKSFAHLKTVNGHQCQTYREACQLLGLLENDSHWDLTLADSVVSSNAYQIRTLFAIIITTCFPSQPIQLWNKFKDDICEDILHRLRIQTNNPDIQITDEIYNEGLILIEDQCLTIANKLLIEVGMIAPNRSMHDAFNQELNRELQYNVDTLQEFVRNNVPLLNEQQKQVYETLMQAVDNNTGGLFFLDAPGGTGKTFVISLILATIRSRCDIALALASSGIAATLLDGGQHFNETDNLRTRITLRFSIYFGFTFIYLEKFN
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
EC Number
3.6.4.12
Pubmed
Proteomes
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Ontologies
GO
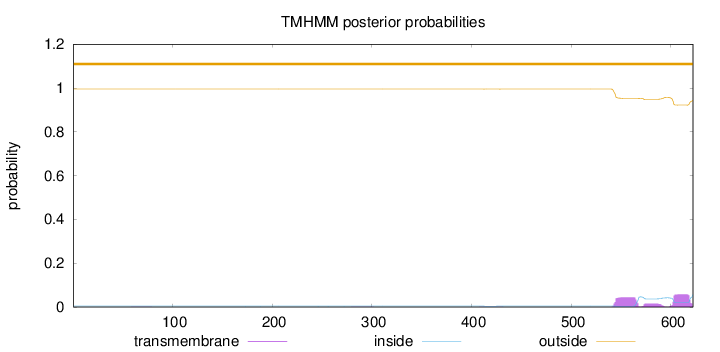
Topology
Length:
622
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.32881
Exp number, first 60 AAs:
0.00138
Total prob of N-in:
0.00462
outside
1 - 622
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
29.519549
CSRT
0
Interpretation
Uncertain
