Gene
KWMTBOMO15741
Pre Gene Modal
BGIBMGA002326
Annotation
PREDICTED:_octopamine_receptor_beta-3R-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.828
Sequence
CDS
ATGGGAGATATAGATGGTGTATTCCGGAGCGAGGTAGAAGCGCGAGGGCGCGGTACGGAGTCAGCGTTTCTCGACGGGCTGCATTCAACGCTGGCGCCTTCTGCCGCGCCCGACCACGACTGGCAGCATACTTTCATCCTCATCATCAAAGGCCTCATCTTCTCCACAATAATCCTCGGCGCTTTATTTGGCAACGCTCTCGTTATCATATCAGTACATCGACACAGGAAGCTCAGAGTTCTCACTAACTACTACGTTGTTAGTCTTGCTGTGGCTGATATGTTAGTCGCCTTATGCGCAATGACCTTTAACGCCAGCTCCGAATTAACAGACGCCTGGTTGTTTGGTCCGTTAGCCTGTGATATCTTTAATTCTCTTGACGTTTACTTCTCCAGTGCTTCTATACTTCATCTGTGCTGCATATCTGTTGATAGATATTACGCTATAGTGCGGCCCCTCGAGTACCCGGTTACCATGACTCATAGGACTGTTTGCTTCATGCTCGCAAACGTTTGGCTGTGGCCCGCGTTCATTAGTTTCGTGCCCATATTCAAGGGTTGGTATACGACTGAACAACACCGCCAATATAGGAAGGACCATCCTGATGTGTGCATTTTCAAAGTCAATATGGTATATGCTATAGTATCTTCGAGCGTTTCATTCTGGATTCCGAGTTTGGTCATGATTTCGATGTACTGCCGAATATTCCGGGAAGCGATCAGACAAAGAGAAGCCTTAACGAGAACTTCGTCAAATATTCTATTGAACTCTGTACATATAAAGCACACGTCACATGCTGCTCATCATTCCCATTATCTGCACCCTGATAGAAGACCTTCAGATATAAGCCCAAACTACAGCACATTCACTGAAGTGGGTCCTGAAGAAACTGAGTTTGGCGAAGTTGTTATGTCTTTGGGAGATATCTGTCCTAGTAAAGAGGCCAGCCCGAGAAAATCTGTGAACGTTAGCTGTGATACTGCTAGTTCGGGGTACTGTTCCGGAGGATCGAACAAGAGATCTTCAAGTGGTCAACCACCTTATGGATGGAGACCAAGTTGTTCTTCCGCGATTGCTACAAGAGATCTTGCAAAAGCTGCCACAGAGCTGAATGCGCAAGGGGCAACGCTTCGTGCCGCTGGGAAGTCGTGGCGTGCTGAGCACAAAGCAGCCCGTACACTTGGCATCATTGTAGGAGCATTCCTGCTGTGCTGGCTACCATTTTTTCTTTGGTACGTTACGACGAACGTGTGCGGGGAGCCTTGCGAGACGCCTTCGGTTGTGGTCGGTGTGCTTTTCTGGATCGGCTATTTCAATTCGGCCTTGAATCCACTGATTTACGCGTATTTCAATCGAGATTTCCGGGACGCGTTTAAGAACACGCTGATGTGCGCCCTGCCCTGTTTGTTCGGTTGTTGGAAGGACACGGGTACGGCCCAATTTGTGTAG
Protein
MGDIDGVFRSEVEARGRGTESAFLDGLHSTLAPSAAPDHDWQHTFILIIKGLIFSTIILGALFGNALVIISVHRHRKLRVLTNYYVVSLAVADMLVALCAMTFNASSELTDAWLFGPLACDIFNSLDVYFSSASILHLCCISVDRYYAIVRPLEYPVTMTHRTVCFMLANVWLWPAFISFVPIFKGWYTTEQHRQYRKDHPDVCIFKVNMVYAIVSSSVSFWIPSLVMISMYCRIFREAIRQREALTRTSSNILLNSVHIKHTSHAAHHSHYLHPDRRPSDISPNYSTFTEVGPEETEFGEVVMSLGDICPSKEASPRKSVNVSCDTASSGYCSGGSNKRSSSGQPPYGWRPSCSSAIATRDLAKAATELNAQGATLRAAGKSWRAEHKAARTLGIIVGAFLLCWLPFFLWYVTTNVCGEPCETPSVVVGVLFWIGYFNSALNPLIYAYFNRDFRDAFKNTLMCALPCLFGCWKDTGTAQFV
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
A0A3S2M7G8
A0A0N1PGA0
H9IYJ2
A0A194QJT9
A0A212F9X2
A0A2H1WUU7
+ More
A0A0L7LSF4 X2KWM1 A0A2J7PYM3 A0A2P8Z628 A0A195BMN4 A0A151X6M7 A0A067RFZ0 D5JAN8 A0A164VF06 A0A0P5GWC7 A0A0P5D8L9 A0A0P5AI88 A0A0N8D376 A0A0P5DSZ0 E0VX90 A0A154P6W6 A0A0P5WF74 A0A0P5BGL3 A0A0P6BG37 A0A1B3P9B4 A0A0P5DHM6 U4UT71 N6TLS1 A0A1B0GLH6 D6WZ38 A0A195BLI6 A0A0P5E0G1 A0A0P6IHB3 A0A067R718 A0A195CML9 A0A1B3P985 D7P7W9 A0A3R7PH95 A0A221LCB7 A0A0N1IPC8
A0A0L7LSF4 X2KWM1 A0A2J7PYM3 A0A2P8Z628 A0A195BMN4 A0A151X6M7 A0A067RFZ0 D5JAN8 A0A164VF06 A0A0P5GWC7 A0A0P5D8L9 A0A0P5AI88 A0A0N8D376 A0A0P5DSZ0 E0VX90 A0A154P6W6 A0A0P5WF74 A0A0P5BGL3 A0A0P6BG37 A0A1B3P9B4 A0A0P5DHM6 U4UT71 N6TLS1 A0A1B0GLH6 D6WZ38 A0A195BLI6 A0A0P5E0G1 A0A0P6IHB3 A0A067R718 A0A195CML9 A0A1B3P985 D7P7W9 A0A3R7PH95 A0A221LCB7 A0A0N1IPC8
Pubmed
EMBL
RSAL01000019
RVE52789.1
KQ460794
KPJ12181.1
BABH01020780
BABH01020781
+ More
KQ458761 KPJ05180.1 AGBW02009547 OWR50545.1 ODYU01011181 SOQ56762.1 JTDY01000237 KOB78121.1 KJ152560 AHN85843.1 NEVH01020342 PNF21441.1 PYGN01000178 PSN51955.1 KQ976440 KYM86546.1 KQ982480 KYQ56017.1 KK852660 KDR19123.1 GU237483 ADD91575.1 LRGB01001361 KZS12256.1 GDIQ01236042 JAK15683.1 GDIP01161383 JAJ62019.1 GDIP01198475 JAJ24927.1 GDIP01073239 JAM30476.1 GDIP01152091 JAJ71311.1 DS235830 EEB17996.1 KQ434829 KZC07669.1 GDIP01087549 JAM16166.1 GDIP01185077 JAJ38325.1 GDIP01016528 JAM87187.1 KU710371 AOG14367.1 GDIP01156094 JAJ67308.1 KI207726 KI208934 KI210288 ERL95728.1 ERL95954.1 ERL96228.1 APGK01000149 KB741407 ENN70180.1 AJWK01035015 AJWK01035016 AJWK01035017 AJWK01035018 AJWK01035019 KQ971372 EFA10770.2 KYM86550.1 GDIP01153092 JAJ70310.1 GDIQ01005074 JAN89663.1 KDR19125.1 KQ977580 KYN01722.1 KU710372 AOG14368.1 GU074419 ADI56272.1 QCYY01002935 ROT66429.1 KX578903 ASM90736.1 KQ460620 KPJ13529.1
KQ458761 KPJ05180.1 AGBW02009547 OWR50545.1 ODYU01011181 SOQ56762.1 JTDY01000237 KOB78121.1 KJ152560 AHN85843.1 NEVH01020342 PNF21441.1 PYGN01000178 PSN51955.1 KQ976440 KYM86546.1 KQ982480 KYQ56017.1 KK852660 KDR19123.1 GU237483 ADD91575.1 LRGB01001361 KZS12256.1 GDIQ01236042 JAK15683.1 GDIP01161383 JAJ62019.1 GDIP01198475 JAJ24927.1 GDIP01073239 JAM30476.1 GDIP01152091 JAJ71311.1 DS235830 EEB17996.1 KQ434829 KZC07669.1 GDIP01087549 JAM16166.1 GDIP01185077 JAJ38325.1 GDIP01016528 JAM87187.1 KU710371 AOG14367.1 GDIP01156094 JAJ67308.1 KI207726 KI208934 KI210288 ERL95728.1 ERL95954.1 ERL96228.1 APGK01000149 KB741407 ENN70180.1 AJWK01035015 AJWK01035016 AJWK01035017 AJWK01035018 AJWK01035019 KQ971372 EFA10770.2 KYM86550.1 GDIP01153092 JAJ70310.1 GDIQ01005074 JAN89663.1 KDR19125.1 KQ977580 KYN01722.1 KU710372 AOG14368.1 GU074419 ADI56272.1 QCYY01002935 ROT66429.1 KX578903 ASM90736.1 KQ460620 KPJ13529.1
Proteomes
PRIDE
Pfam
PF00001 7tm_1
Interpro
ProteinModelPortal
A0A3S2M7G8
A0A0N1PGA0
H9IYJ2
A0A194QJT9
A0A212F9X2
A0A2H1WUU7
+ More
A0A0L7LSF4 X2KWM1 A0A2J7PYM3 A0A2P8Z628 A0A195BMN4 A0A151X6M7 A0A067RFZ0 D5JAN8 A0A164VF06 A0A0P5GWC7 A0A0P5D8L9 A0A0P5AI88 A0A0N8D376 A0A0P5DSZ0 E0VX90 A0A154P6W6 A0A0P5WF74 A0A0P5BGL3 A0A0P6BG37 A0A1B3P9B4 A0A0P5DHM6 U4UT71 N6TLS1 A0A1B0GLH6 D6WZ38 A0A195BLI6 A0A0P5E0G1 A0A0P6IHB3 A0A067R718 A0A195CML9 A0A1B3P985 D7P7W9 A0A3R7PH95 A0A221LCB7 A0A0N1IPC8
A0A0L7LSF4 X2KWM1 A0A2J7PYM3 A0A2P8Z628 A0A195BMN4 A0A151X6M7 A0A067RFZ0 D5JAN8 A0A164VF06 A0A0P5GWC7 A0A0P5D8L9 A0A0P5AI88 A0A0N8D376 A0A0P5DSZ0 E0VX90 A0A154P6W6 A0A0P5WF74 A0A0P5BGL3 A0A0P6BG37 A0A1B3P9B4 A0A0P5DHM6 U4UT71 N6TLS1 A0A1B0GLH6 D6WZ38 A0A195BLI6 A0A0P5E0G1 A0A0P6IHB3 A0A067R718 A0A195CML9 A0A1B3P985 D7P7W9 A0A3R7PH95 A0A221LCB7 A0A0N1IPC8
PDB
2R4R
E-value=2.41016e-39,
Score=408
Ontologies
GO
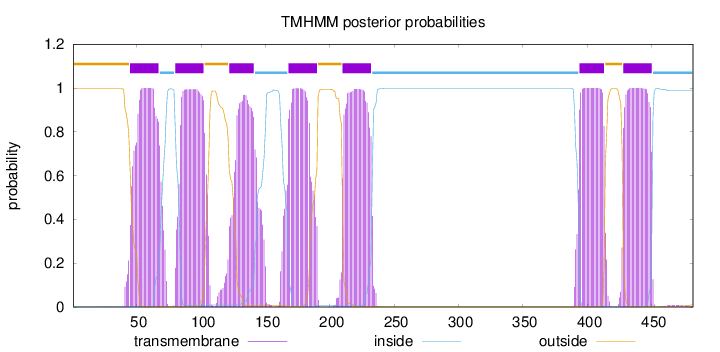
Topology
Subcellular location
Cell membrane
Length:
482
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
155.48441
Exp number, first 60 AAs:
14.1593
Total prob of N-in:
0.00340
POSSIBLE N-term signal
sequence
outside
1 - 44
TMhelix
45 - 67
inside
68 - 79
TMhelix
80 - 102
outside
103 - 121
TMhelix
122 - 141
inside
142 - 167
TMhelix
168 - 190
outside
191 - 209
TMhelix
210 - 232
inside
233 - 393
TMhelix
394 - 413
outside
414 - 427
TMhelix
428 - 450
inside
451 - 482
Population Genetic Test Statistics
Pi
225.714265
Theta
169.026301
Tajima's D
1.163272
CLR
0.24445
CSRT
0.709514524273786
Interpretation
Uncertain
