Gene
KWMTBOMO15740
Pre Gene Modal
BGIBMGA010159
Annotation
PREDICTED:_dynamin-like_120_kDa_protein?_mitochondrial_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.673
Sequence
CDS
ATGGCAGAACTCATTGAAATTCAATCCATGATTTATGACAACATTGTCAAACTTAGCATCAACATAAAAAAGGATAGTGCGGATAGAAAGACGGCTGATTATTTTAAGAGAAGGTTGAGTACTTTAGAGAGCTACTGGCAAGAATGCCAATTTAACCATGACCGTATACAAACCGAAATAAGTAAGACTCATCCTTATTTCATCGAACAACAGTATGAAAAAATATATCAAACGTATGAGGCAACCAAGAATATGATTAACGAGCAGTATCAGCGACTAGTAGAACAAGTAGGCATTCTTCAAGAAGGGGAGGAAGCTGGTGCCCGTTCCCGGACGCCAGCAGGAGCGAAGTCAAAACAAATAAATGAAGAAACAAGAACCAGTGGGCAAAGCCAACAGGTTGAACAAATGAAAAGAAGCCAAGGCACAAACAGTAAATTGGATGATTTCTTAAAAAGACAAAATGCTAATTTTAAAGCCTTTATACGAACAACATCAAACATAGACGTAGATTTGATTGCTGATAAATGGGAATTTGAAGACTCTTTAAAAACACTACACGAACGATGGGCTGCTATCGATAGCCTTCACTGGGAAATCGAATCCGAGATGGACGGAGAAAATGAAACATACGCCGCCATGTACGATAGATACGAGAAGAAATTTAATACTCTCAAGAAAACATTGAACACAAAGATGTGGTCGGTAGCTCATAGATCGAATGCTACTCCACAATTGGAAATACCTGTCTTCAGCGGAAATTATAATCAGTGGGTTTCTTTCAAAGATTTATTCACGGAAGCTGTTCACAACAATCCATCTCTATCTAACAGTCAAAAAATGCAACACCTGAAAACTCCGGTGTCAACTGAACAAAACATTGCGAAACAACGCATAACACGAAATGAGAATGTTTCAAAAACCTCTAATGCTTCGCTAAATGAGAGTTCAGAAGTTTTATTAGCTACTGCTTTGGTGAAGGTGAAAGCGGCTGATGGAACCTATTACAACATGCGAGCTTTGTTAGATCAAGGCTCGCAAATATCTTTGATAACTGAAGCAGCAGCCCAAACATTAGGATTAAAGCGTCAAAGATGCAAAGGGGTTATATTTGGTGTAGGCCAAAACGAAAACAGCTGCAAAGGAAAGGTAATCATAGAATGCTCCTCAATTAACAATGACTTCAATTTCAAACTAGAGGCACTCATTATGAACAATTTAATAAAAAGTTTACCAAACAAAACATTTTCTAAGCCAGCGTGGTCATATTTGAATAATATCAATTTAGCTGATCCCGAGTTTTACAAGAGTCGGCCAGTCGATCTACTGTTCGGTGCGGATATCTACGCTAACATTATTCTGGGAGGAGTGATTAAAGGAGAAACCTTACAACAACCCATGGCTCAACAATCACAGTTAGGATGGTTTTTATGCGGAAGTATTAAAACTTATCATTGTAATGTGGCTCTCAATAACGTGGAAGATATACATAAATTTTGGGAGGTGGAAGAAATCAACGAATCGACGAATATTTCCGGAGAAGATCAGGAATGTATCGACTTTTACTGCTCAACAACAAAACGCATTGAAAATGGTCGATATGAAGTGAGATTGCCTCTAAAGCAAGATTTAAAACATAATTTAGGAATGAAGGCATTCGAAAAGGCGACAACACATTTAGATAAATCATGCAAAGTCCTGGAAGATGGTCAGTGTAGCATAATAGTCGATCAATTGATCCACGAATACAAAGAATTAACTTACTATAATGAGCTACTTATGAATCAACATTTGAATGAGCACTACAGACAGCGTCGTGGTTTTATCAACGGTGTCGGATATCTTGCTAACTCTCTGTTCGGAGTATTAGATGAACATTTCGCAGAAAAATACTCGCAAGACATAGAGCTCGTAAAACATAATGAACAGTACTTAGTGAGTTTATGGAAAAACCAAACTTCGATCGTCGAATCTGAATACAACTTACTAAAAAGAACAGAAGAAGTCATGTCTAAGCAACATAAGACTATTAATAAACACATAACTAACCTAGAAAACATGTCAAATCGACTTCAACAGCAAGTAAACAACATATCGATTCTACAAGAATTTACCTTAATGTCCATTATTACAACTAATTTATTGCACAATTTGAAAGGGTCGCAAAATATATTCTTAGACATTTTAACTAACATACATCATGGTCAATTCAACATTCATTTACTTACTCCAGAACAACTTCAAAAGGAACTAAACATTATCACCGGATATTTACCGGAAGACACAATTCTTCCCGTAGAACCGCAGAATATTAAGAATTTGTATCCTCTATTAAACATAAAGGCTCGTATTACTCTACAATATTTCTTATTCGAGATAACATTTCCACTCATTTATCAAGATCGATTCAAACTTTATAAAGTACATTCTATTCCTCATCAAAGCGGAAATATGACTATTGAAGTAGTTCCGGCATACGAATACATTGCTACCGACTTGAAAAGAGATTCCTTTCTTGAACTCACCACCGAAGAAGTCTTATCCTGTAATCAACAAGAAGAAGCTTACTACTGCAAGCTTAGAACGCCGATTCTTCATTTACGACCGGAGACTAAGTTTTGTAAGATTAAACTTAATAGCAACGTATGCAAATTAAAGTATTCTAATTGTGAAACCAAAATAATAGCACTAACTCATCCCTCTTTATATTTATATCACTGCTGTGGCACCTGCGAAGTAAAGGTCATGTGCGGAGAACATATTGCCTTGAAACAGTTGAATAACACTGGTCGTCTGTATCTAGAGCCAGGTTGCCTCATAAAGGGATCAGATTATACACTTCATATCCCTAAAACAAACAACAATAAGCTGGAGATTCAGTCTAACATTTACGTTCCAATAATAGATCCCATAAATAATATCATAAACAGCTCTCTTCTCAACACTGAGAACCTGAATGTCAGCGAGTCTAAAGATTTGCAGAACGCTCTCCAGAAACTACACGTGCGTATTGAAGATCTAGAAAACTCCCCGCCAGTATTGGAGACTTCATTAACATCTCACGATATTCATCAGTATGTGATAACATACATCATCGTCATCGTCATCGTGCTGGCCACCGGCGTGCTACTCTGGCGACGACGACGGTTGAGTGCGAACCGGGCGATCATCTCTGACGGCATCGAGCTTCAACAGTATAGTGAATGTGAAAATGTAAATAGTGCTAAGGCAGGGTTTAGTGCGCGACCTTCTGTGGTGGACCCTTCGCCTGGTGATGTGAACATTTATGCCAAAGTGTCAAACAAAGCGTGCTCTCCAATACCAAGAAAACCTATATGGACCCTAAGTTCTTAA
Protein
MAELIEIQSMIYDNIVKLSINIKKDSADRKTADYFKRRLSTLESYWQECQFNHDRIQTEISKTHPYFIEQQYEKIYQTYEATKNMINEQYQRLVEQVGILQEGEEAGARSRTPAGAKSKQINEETRTSGQSQQVEQMKRSQGTNSKLDDFLKRQNANFKAFIRTTSNIDVDLIADKWEFEDSLKTLHERWAAIDSLHWEIESEMDGENETYAAMYDRYEKKFNTLKKTLNTKMWSVAHRSNATPQLEIPVFSGNYNQWVSFKDLFTEAVHNNPSLSNSQKMQHLKTPVSTEQNIAKQRITRNENVSKTSNASLNESSEVLLATALVKVKAADGTYYNMRALLDQGSQISLITEAAAQTLGLKRQRCKGVIFGVGQNENSCKGKVIIECSSINNDFNFKLEALIMNNLIKSLPNKTFSKPAWSYLNNINLADPEFYKSRPVDLLFGADIYANIILGGVIKGETLQQPMAQQSQLGWFLCGSIKTYHCNVALNNVEDIHKFWEVEEINESTNISGEDQECIDFYCSTTKRIENGRYEVRLPLKQDLKHNLGMKAFEKATTHLDKSCKVLEDGQCSIIVDQLIHEYKELTYYNELLMNQHLNEHYRQRRGFINGVGYLANSLFGVLDEHFAEKYSQDIELVKHNEQYLVSLWKNQTSIVESEYNLLKRTEEVMSKQHKTINKHITNLENMSNRLQQQVNNISILQEFTLMSIITTNLLHNLKGSQNIFLDILTNIHHGQFNIHLLTPEQLQKELNIITGYLPEDTILPVEPQNIKNLYPLLNIKARITLQYFLFEITFPLIYQDRFKLYKVHSIPHQSGNMTIEVVPAYEYIATDLKRDSFLELTTEEVLSCNQQEEAYYCKLRTPILHLRPETKFCKIKLNSNVCKLKYSNCETKIIALTHPSLYLYHCCGTCEVKVMCGEHIALKQLNNTGRLYLEPGCLIKGSDYTLHIPKTNNNKLEIQSNIYVPIIDPINNIINSSLLNTENLNVSESKDLQNALQKLHVRIEDLENSPPVLETSLTSHDIHQYVITYIIVIVIVLATGVLLWRRRRLSANRAIISDGIELQQYSECENVNSAKAGFSARPSVVDPSPGDVNIYAKVSNKACSPIPRKPIWTLSS
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
Topology
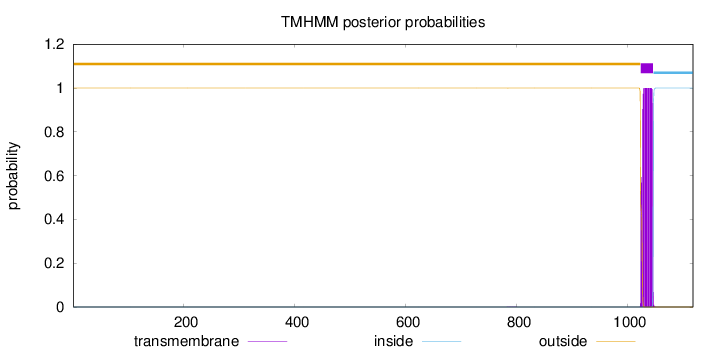
Length:
1117
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.59555
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 1022
TMhelix
1023 - 1045
inside
1046 - 1117
Population Genetic Test Statistics
Pi
187.319863
Theta
155.265029
Tajima's D
0.500625
CLR
39.915057
CSRT
0.515024248787561
Interpretation
Uncertain
