Gene
KWMTBOMO15735
Pre Gene Modal
BGIBMGA002324
Annotation
PREDICTED:_sodium-coupled_monocarboxylate_transporter_1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.973
Sequence
CDS
ATGAAGCCAGGATGGTTAGCATGGGAGTGTATAGTGTTCGCTATGTTTGTGTGTTCAACCTCCTGCGCCCCGCTGTGGCGTAGGCGAAGAGATGCTGCGGCCGGTGGCAGTGCCAAGAGAGCATACATCTTCGCGGGCACCGGAGTGTCAACTTTAGCTATGATACTATCGGTGGCTAGAGGAACGCTAGGGGTTCGATCGTTTTTAGGGTTTCCGTCGGAGCAAGTATACCGAGGGTCAGCTATGTGGGAAACGCTTTTTGGCATGGTGCTAGCGTTTCCCATAGTTTGCTGGATCTTCGTCCCGGTCTATTACAGGCTATCAACTAATTCCGTTTATGAATATTTGGAGATGCGTTTCGGTTCGATTTGGGTTCGGCGCATAGCCGCAGCAACTTTTCTCCTCCGGCAAGTTTTGAATCTGGCCGTGACGGTCTACACTCCAACAGTGGCCCTTCATGCTGTACTGGCACTACCACACTGGGCGTCAGCAGCTGCACTGACCGTCGTTAGCATCGTTTTCAATTTACTCGGCGGGCTTGCAGCTGCTATTAGAGCTGATGTCATTCAGACTTTGACCATGATACTCGTGAGTGGAGCTTTCATAGTTCAAGCGACAGTGAAAGCTGGTGGTCCGGCAACGGTGTTTAATGGCAATCTAGAAGGCGGGAGACTTAAGTTTTTCCAATTCACGTGGGACCCGACCGTTAGAGTGGACACACTATCCGCCATTATCGGGCAGCTATTCATGTCCATATCCATCTATGGATGCCAGCAGACTTTTGTCCAGCGTTACTGCTCCATGTCCAGCGAATCCAGGGTCAAGAAAACTCTCCTGGCCAACGTTCCTGCCGTTTCGGTGCTCTTCAGTTTGTCTTGGATCGTAGGCATGGCGCTATATGCTGTGTACAAATACTGTGATCCACTGGCCACAAGCAAAATTTCGGCCCCCGATGAAATCCTGCCATTTTACGTACAAGACCAATTTGGTTTTCTACCAGGGATGTTGGGACTGTTTCTGGGCAGTCTTTTCAATGGCGCTTTGAGTTTCCTGGTATCAAACGTTAATTCCTTATCCACAGTCACATGGGAGGATTTTGTATCCGCTGCTCCGTCTTTTAAAGGAATCAGTGATAAGCAACAGCTTACTATCATTAAAATCATTGGGGTAATCTATTCGATCGTCATAATGTGCTTGTCTCTCTGCGTGGGCATGGCTGGTGGCGTAGTCGAAGGCTCCCTACTAGTGACGTCAGCCACGTCCGGAGCACTCTTAGGAGTGTTTGTCCTGGCAGCTCTGTGTCCGGCTGCTAACGGTACGGGCGCGGTATGGGGAATGATCGCTTCGCACATTATAACGACCTGGATGGCTGTTGGAAGGCTCTTGTATGTGAAAAAGAATATAGCCATGCTGCCTACTTCGGTCGAGGGCTGCACGAACACAACGCTCAATTACCTGAGTCGGCCCGTGGGCATCAACCAGACCCTGGCCAAACTAGCACCCCTGCTCCAATCTATGAACATCAGCCTAACAGAAATACGTCCACCGGAATCGTTCATTATGTCCAACCTTCACAAGATGTATTCAATCTCGTATATGTGGTACGCTGTTATCGGCACAGTCACCTGCGTGACGATAGGCGTGATCATCAGCCTCCTGACTTGGAAGGAAACGGACAGATACGACGAGAAGCTCCTACACCCGCTTGTAGCGAAACTGAGCAGGAAACTACCGGGCAGGATTCGTACGTTACCGATCGAGAAGTCTAATATAGAGGTGAAAGAGGACAAGGAAAGTTCAGAGAAGACTGACCAGACTGTAGTGGATGCGAAGCCTACGGTTTTCGCCACTACCAGCAGCAGACTGTTTGAGGCTTACGAAGTCAGAAGATCGCCTTCTCCTGCTCGGACGCGTTTATGA
Protein
MKPGWLAWECIVFAMFVCSTSCAPLWRRRRDAAAGGSAKRAYIFAGTGVSTLAMILSVARGTLGVRSFLGFPSEQVYRGSAMWETLFGMVLAFPIVCWIFVPVYYRLSTNSVYEYLEMRFGSIWVRRIAAATFLLRQVLNLAVTVYTPTVALHAVLALPHWASAAALTVVSIVFNLLGGLAAAIRADVIQTLTMILVSGAFIVQATVKAGGPATVFNGNLEGGRLKFFQFTWDPTVRVDTLSAIIGQLFMSISIYGCQQTFVQRYCSMSSESRVKKTLLANVPAVSVLFSLSWIVGMALYAVYKYCDPLATSKISAPDEILPFYVQDQFGFLPGMLGLFLGSLFNGALSFLVSNVNSLSTVTWEDFVSAAPSFKGISDKQQLTIIKIIGVIYSIVIMCLSLCVGMAGGVVEGSLLVTSATSGALLGVFVLAALCPAANGTGAVWGMIASHIITTWMAVGRLLYVKKNIAMLPTSVEGCTNTTLNYLSRPVGINQTLAKLAPLLQSMNISLTEIRPPESFIMSNLHKMYSISYMWYAVIGTVTCVTIGVIISLLTWKETDRYDEKLLHPLVAKLSRKLPGRIRTLPIEKSNIEVKEDKESSEKTDQTVVDAKPTVFATTSSRLFEAYEVRRSPSPARTRL
Summary
Similarity
Belongs to the sodium:solute symporter (SSF) (TC 2.A.21) family.
Uniprot
A0A2A4K6R0
A0A2W1BKY4
A0A194QJT4
H9IYJ0
A0A212EH53
A0A2H1WF29
+ More
S4PG51 A0A3S2N668 A0A1Q3FPC8 A0A1Q3FP13 B0W5X1 A0A2M4BGH3 A0A2M4BH38 A0A336LQ40 A0A182R603 A0A2M3YYV2 A0A2M3YYR5 A0A084W5H1 A0A182M285 A0A0T6BHG4 A0A182NS24 A0A2M4A1K2 W5J713 A0A2M4A165 D6X0X6 A0A182J792 A0A182YB68 A0A182VZE0 Q17HV6 A0A182V813 A0A182FQ25 A0A182L9T5 Q7PXX1 A0A182HLE9 A0A182WSJ7 A0A0C9QWN5 A0A1Y1LEB8 E2C717 A0A182TVZ2 A0A182PGL8 U4UEV5 A0A0J7L2S0 F4X723 A0A182GFW0 A0A232FEK9 A0A182K018 A0A1J1HNB9 E9IVB9 A0A182QU24 A0A195DK21 A0A195FJ08 K7IU91 A0A088AGB8 A0A151XHC6 E2A500 A0A2A3EPE9 A0A1W4X697 A0A026WHY4 A0A1W4UAZ8 A0A2J7RFS4 A0A3B0KG79 B4G3Q0 A0A195C0N7 A0A3B0JJX3 B3MSQ7 Q298F5 B4MC07 A0A0P8XE45 A0A0K8U1G8 A0A0M3QXJ9 A0A195B0I7 A0A0M9A9W7 A0A2M3YZ90 A0A0A1WRY5 B4PM18 B4K597 Q7K3Q2 A0A1L8DES3 B3P3A8 W8BY77 A0A158NDB7 B4QTY8 B4I250 A0A1L8DEY9 B4NFZ7 A0A1B6G4A6 A0A1B0CBU0 A0A1S4F253 A0A1B6HXP6 A0A182G6Y5 A0A1B6IMK7 Q17HV7 A0A1B0GAB6 A0A1B6CB84 B4JV34 A0A0L7RJF8 A0A154PI07 A0A310S9B8 A0A0A9ZGA3 B0W5X4
S4PG51 A0A3S2N668 A0A1Q3FPC8 A0A1Q3FP13 B0W5X1 A0A2M4BGH3 A0A2M4BH38 A0A336LQ40 A0A182R603 A0A2M3YYV2 A0A2M3YYR5 A0A084W5H1 A0A182M285 A0A0T6BHG4 A0A182NS24 A0A2M4A1K2 W5J713 A0A2M4A165 D6X0X6 A0A182J792 A0A182YB68 A0A182VZE0 Q17HV6 A0A182V813 A0A182FQ25 A0A182L9T5 Q7PXX1 A0A182HLE9 A0A182WSJ7 A0A0C9QWN5 A0A1Y1LEB8 E2C717 A0A182TVZ2 A0A182PGL8 U4UEV5 A0A0J7L2S0 F4X723 A0A182GFW0 A0A232FEK9 A0A182K018 A0A1J1HNB9 E9IVB9 A0A182QU24 A0A195DK21 A0A195FJ08 K7IU91 A0A088AGB8 A0A151XHC6 E2A500 A0A2A3EPE9 A0A1W4X697 A0A026WHY4 A0A1W4UAZ8 A0A2J7RFS4 A0A3B0KG79 B4G3Q0 A0A195C0N7 A0A3B0JJX3 B3MSQ7 Q298F5 B4MC07 A0A0P8XE45 A0A0K8U1G8 A0A0M3QXJ9 A0A195B0I7 A0A0M9A9W7 A0A2M3YZ90 A0A0A1WRY5 B4PM18 B4K597 Q7K3Q2 A0A1L8DES3 B3P3A8 W8BY77 A0A158NDB7 B4QTY8 B4I250 A0A1L8DEY9 B4NFZ7 A0A1B6G4A6 A0A1B0CBU0 A0A1S4F253 A0A1B6HXP6 A0A182G6Y5 A0A1B6IMK7 Q17HV7 A0A1B0GAB6 A0A1B6CB84 B4JV34 A0A0L7RJF8 A0A154PI07 A0A310S9B8 A0A0A9ZGA3 B0W5X4
Pubmed
28756777
26354079
19121390
22118469
23622113
24438588
+ More
20920257 23761445 18362917 19820115 25244985 17510324 20966253 12364791 28004739 20798317 23537049 21719571 26483478 28648823 21282665 20075255 24508170 30249741 17994087 18057021 15632085 23185243 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 21347285 25401762 26823975
20920257 23761445 18362917 19820115 25244985 17510324 20966253 12364791 28004739 20798317 23537049 21719571 26483478 28648823 21282665 20075255 24508170 30249741 17994087 18057021 15632085 23185243 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 21347285 25401762 26823975
EMBL
NWSH01000072
PCG79955.1
KZ150018
PZC74941.1
KQ458761
KPJ05175.1
+ More
BABH01020722 AGBW02014970 OWR40800.1 ODYU01008262 SOQ51695.1 GAIX01003782 JAA88778.1 RSAL01000325 RVE42526.1 GFDL01005707 JAV29338.1 GFDL01005771 JAV29274.1 DS231845 EDS35958.1 GGFJ01002976 MBW52117.1 GGFJ01002977 MBW52118.1 UFQT01000111 SSX20196.1 GGFM01000692 MBW21443.1 GGFM01000658 MBW21409.1 ATLV01020636 KE525303 KFB45465.1 AXCM01000994 LJIG01000235 KRT86705.1 GGFK01001197 MBW34518.1 ADMH02002100 ETN59188.1 GGFK01001202 MBW34523.1 KQ971372 EFA09536.1 CH477245 EAT46233.1 AAAB01008987 EAA00973.4 APCN01000896 GBYB01000118 GBYB01000119 GBYB01000120 GBYB01000121 JAG69885.1 JAG69886.1 JAG69887.1 JAG69888.1 GEZM01061795 GEZM01061794 GEZM01061793 JAV70315.1 GL453255 EFN76289.1 KB632063 ERL88445.1 LBMM01001017 KMQ97062.1 GL888828 EGI57635.1 JXUM01060500 KQ562105 KXJ76677.1 NNAY01000338 OXU29092.1 CVRI01000010 CRK89018.1 GL766194 EFZ15458.1 AXCN02000264 KQ980800 KYN12834.1 KQ981560 KYN39969.1 AAZX01006375 KQ982130 KYQ59814.1 GL436778 EFN71487.1 KZ288200 PBC33580.1 KK107199 QOIP01000009 EZA55610.1 RLU18719.1 NEVH01004410 PNF39682.1 OUUW01000007 SPP82648.1 CH479179 EDW24371.1 KQ978379 KYM94412.1 SPP82647.1 CH902623 EDV30297.1 KPU72780.1 KPU72781.1 CM000070 EAL28000.2 KRT00372.1 CH940656 EDW58628.1 KRF78368.1 KRF78369.1 KRF78370.1 KPU72782.1 GDHF01031968 GDHF01018179 GDHF01010176 JAI20346.1 JAI34135.1 JAI42138.1 CP012526 ALC46063.1 KQ976690 KYM77993.1 KQ435706 KOX80190.1 GGFM01000815 MBW21566.1 GBXI01013089 JAD01203.1 CM000160 EDW95950.1 KRK02808.1 CH933806 EDW15097.1 KRG01361.1 KRG01362.1 KRG01363.1 KRG01364.1 KRG01365.1 KRG01366.1 KRG01367.1 AE014297 AY058425 AAF55596.1 AAL13654.1 GFDF01009138 JAV04946.1 CH954181 EDV48560.1 GAMC01008295 GAMC01008294 GAMC01008293 JAB98261.1 ADTU01012549 ADTU01012550 CM000364 EDX12411.1 CH480820 EDW54607.1 GFDF01009137 JAV04947.1 CH964251 EDW83214.2 GECZ01012499 JAS57270.1 AJWK01005824 AJWK01005825 AJWK01005826 AJWK01005827 AJWK01005828 GECU01036461 GECU01028291 GECU01012342 GECU01010967 JAS71245.1 JAS79415.1 JAS95364.1 JAS96739.1 JXUM01149218 KQ570728 KXJ68314.1 GECU01032882 GECU01019562 GECU01014578 GECU01002370 JAS74824.1 JAS88144.1 JAS93128.1 JAT05337.1 EAT46232.1 CCAG010002699 GEDC01026699 GEDC01009522 JAS10599.1 JAS27776.1 CH916374 EDV91354.1 KQ414581 KOC70928.1 KQ434902 KZC11124.1 KQ763446 OAD55073.1 GBHO01002604 GBHO01002603 GDHC01015740 GDHC01011490 GDHC01007398 JAG41000.1 JAG41001.1 JAQ02889.1 JAQ07139.1 JAQ11231.1 EDS35961.1
BABH01020722 AGBW02014970 OWR40800.1 ODYU01008262 SOQ51695.1 GAIX01003782 JAA88778.1 RSAL01000325 RVE42526.1 GFDL01005707 JAV29338.1 GFDL01005771 JAV29274.1 DS231845 EDS35958.1 GGFJ01002976 MBW52117.1 GGFJ01002977 MBW52118.1 UFQT01000111 SSX20196.1 GGFM01000692 MBW21443.1 GGFM01000658 MBW21409.1 ATLV01020636 KE525303 KFB45465.1 AXCM01000994 LJIG01000235 KRT86705.1 GGFK01001197 MBW34518.1 ADMH02002100 ETN59188.1 GGFK01001202 MBW34523.1 KQ971372 EFA09536.1 CH477245 EAT46233.1 AAAB01008987 EAA00973.4 APCN01000896 GBYB01000118 GBYB01000119 GBYB01000120 GBYB01000121 JAG69885.1 JAG69886.1 JAG69887.1 JAG69888.1 GEZM01061795 GEZM01061794 GEZM01061793 JAV70315.1 GL453255 EFN76289.1 KB632063 ERL88445.1 LBMM01001017 KMQ97062.1 GL888828 EGI57635.1 JXUM01060500 KQ562105 KXJ76677.1 NNAY01000338 OXU29092.1 CVRI01000010 CRK89018.1 GL766194 EFZ15458.1 AXCN02000264 KQ980800 KYN12834.1 KQ981560 KYN39969.1 AAZX01006375 KQ982130 KYQ59814.1 GL436778 EFN71487.1 KZ288200 PBC33580.1 KK107199 QOIP01000009 EZA55610.1 RLU18719.1 NEVH01004410 PNF39682.1 OUUW01000007 SPP82648.1 CH479179 EDW24371.1 KQ978379 KYM94412.1 SPP82647.1 CH902623 EDV30297.1 KPU72780.1 KPU72781.1 CM000070 EAL28000.2 KRT00372.1 CH940656 EDW58628.1 KRF78368.1 KRF78369.1 KRF78370.1 KPU72782.1 GDHF01031968 GDHF01018179 GDHF01010176 JAI20346.1 JAI34135.1 JAI42138.1 CP012526 ALC46063.1 KQ976690 KYM77993.1 KQ435706 KOX80190.1 GGFM01000815 MBW21566.1 GBXI01013089 JAD01203.1 CM000160 EDW95950.1 KRK02808.1 CH933806 EDW15097.1 KRG01361.1 KRG01362.1 KRG01363.1 KRG01364.1 KRG01365.1 KRG01366.1 KRG01367.1 AE014297 AY058425 AAF55596.1 AAL13654.1 GFDF01009138 JAV04946.1 CH954181 EDV48560.1 GAMC01008295 GAMC01008294 GAMC01008293 JAB98261.1 ADTU01012549 ADTU01012550 CM000364 EDX12411.1 CH480820 EDW54607.1 GFDF01009137 JAV04947.1 CH964251 EDW83214.2 GECZ01012499 JAS57270.1 AJWK01005824 AJWK01005825 AJWK01005826 AJWK01005827 AJWK01005828 GECU01036461 GECU01028291 GECU01012342 GECU01010967 JAS71245.1 JAS79415.1 JAS95364.1 JAS96739.1 JXUM01149218 KQ570728 KXJ68314.1 GECU01032882 GECU01019562 GECU01014578 GECU01002370 JAS74824.1 JAS88144.1 JAS93128.1 JAT05337.1 EAT46232.1 CCAG010002699 GEDC01026699 GEDC01009522 JAS10599.1 JAS27776.1 CH916374 EDV91354.1 KQ414581 KOC70928.1 KQ434902 KZC11124.1 KQ763446 OAD55073.1 GBHO01002604 GBHO01002603 GDHC01015740 GDHC01011490 GDHC01007398 JAG41000.1 JAG41001.1 JAQ02889.1 JAQ07139.1 JAQ11231.1 EDS35961.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000007151
UP000283053
UP000002320
+ More
UP000075900 UP000030765 UP000075883 UP000075884 UP000000673 UP000007266 UP000075880 UP000076408 UP000075920 UP000008820 UP000075903 UP000069272 UP000075882 UP000007062 UP000075840 UP000076407 UP000008237 UP000075902 UP000075885 UP000030742 UP000036403 UP000007755 UP000069940 UP000249989 UP000215335 UP000075881 UP000183832 UP000075886 UP000078492 UP000078541 UP000002358 UP000005203 UP000075809 UP000000311 UP000242457 UP000192223 UP000053097 UP000279307 UP000192221 UP000235965 UP000268350 UP000008744 UP000078542 UP000007801 UP000001819 UP000008792 UP000092553 UP000078540 UP000053105 UP000002282 UP000009192 UP000000803 UP000008711 UP000005205 UP000000304 UP000001292 UP000007798 UP000092461 UP000092444 UP000001070 UP000053825 UP000076502
UP000075900 UP000030765 UP000075883 UP000075884 UP000000673 UP000007266 UP000075880 UP000076408 UP000075920 UP000008820 UP000075903 UP000069272 UP000075882 UP000007062 UP000075840 UP000076407 UP000008237 UP000075902 UP000075885 UP000030742 UP000036403 UP000007755 UP000069940 UP000249989 UP000215335 UP000075881 UP000183832 UP000075886 UP000078492 UP000078541 UP000002358 UP000005203 UP000075809 UP000000311 UP000242457 UP000192223 UP000053097 UP000279307 UP000192221 UP000235965 UP000268350 UP000008744 UP000078542 UP000007801 UP000001819 UP000008792 UP000092553 UP000078540 UP000053105 UP000002282 UP000009192 UP000000803 UP000008711 UP000005205 UP000000304 UP000001292 UP000007798 UP000092461 UP000092444 UP000001070 UP000053825 UP000076502
Pfam
PF00474 SSF
Interpro
SUPFAM
SSF161093
SSF161093
Gene 3D
ProteinModelPortal
A0A2A4K6R0
A0A2W1BKY4
A0A194QJT4
H9IYJ0
A0A212EH53
A0A2H1WF29
+ More
S4PG51 A0A3S2N668 A0A1Q3FPC8 A0A1Q3FP13 B0W5X1 A0A2M4BGH3 A0A2M4BH38 A0A336LQ40 A0A182R603 A0A2M3YYV2 A0A2M3YYR5 A0A084W5H1 A0A182M285 A0A0T6BHG4 A0A182NS24 A0A2M4A1K2 W5J713 A0A2M4A165 D6X0X6 A0A182J792 A0A182YB68 A0A182VZE0 Q17HV6 A0A182V813 A0A182FQ25 A0A182L9T5 Q7PXX1 A0A182HLE9 A0A182WSJ7 A0A0C9QWN5 A0A1Y1LEB8 E2C717 A0A182TVZ2 A0A182PGL8 U4UEV5 A0A0J7L2S0 F4X723 A0A182GFW0 A0A232FEK9 A0A182K018 A0A1J1HNB9 E9IVB9 A0A182QU24 A0A195DK21 A0A195FJ08 K7IU91 A0A088AGB8 A0A151XHC6 E2A500 A0A2A3EPE9 A0A1W4X697 A0A026WHY4 A0A1W4UAZ8 A0A2J7RFS4 A0A3B0KG79 B4G3Q0 A0A195C0N7 A0A3B0JJX3 B3MSQ7 Q298F5 B4MC07 A0A0P8XE45 A0A0K8U1G8 A0A0M3QXJ9 A0A195B0I7 A0A0M9A9W7 A0A2M3YZ90 A0A0A1WRY5 B4PM18 B4K597 Q7K3Q2 A0A1L8DES3 B3P3A8 W8BY77 A0A158NDB7 B4QTY8 B4I250 A0A1L8DEY9 B4NFZ7 A0A1B6G4A6 A0A1B0CBU0 A0A1S4F253 A0A1B6HXP6 A0A182G6Y5 A0A1B6IMK7 Q17HV7 A0A1B0GAB6 A0A1B6CB84 B4JV34 A0A0L7RJF8 A0A154PI07 A0A310S9B8 A0A0A9ZGA3 B0W5X4
S4PG51 A0A3S2N668 A0A1Q3FPC8 A0A1Q3FP13 B0W5X1 A0A2M4BGH3 A0A2M4BH38 A0A336LQ40 A0A182R603 A0A2M3YYV2 A0A2M3YYR5 A0A084W5H1 A0A182M285 A0A0T6BHG4 A0A182NS24 A0A2M4A1K2 W5J713 A0A2M4A165 D6X0X6 A0A182J792 A0A182YB68 A0A182VZE0 Q17HV6 A0A182V813 A0A182FQ25 A0A182L9T5 Q7PXX1 A0A182HLE9 A0A182WSJ7 A0A0C9QWN5 A0A1Y1LEB8 E2C717 A0A182TVZ2 A0A182PGL8 U4UEV5 A0A0J7L2S0 F4X723 A0A182GFW0 A0A232FEK9 A0A182K018 A0A1J1HNB9 E9IVB9 A0A182QU24 A0A195DK21 A0A195FJ08 K7IU91 A0A088AGB8 A0A151XHC6 E2A500 A0A2A3EPE9 A0A1W4X697 A0A026WHY4 A0A1W4UAZ8 A0A2J7RFS4 A0A3B0KG79 B4G3Q0 A0A195C0N7 A0A3B0JJX3 B3MSQ7 Q298F5 B4MC07 A0A0P8XE45 A0A0K8U1G8 A0A0M3QXJ9 A0A195B0I7 A0A0M9A9W7 A0A2M3YZ90 A0A0A1WRY5 B4PM18 B4K597 Q7K3Q2 A0A1L8DES3 B3P3A8 W8BY77 A0A158NDB7 B4QTY8 B4I250 A0A1L8DEY9 B4NFZ7 A0A1B6G4A6 A0A1B0CBU0 A0A1S4F253 A0A1B6HXP6 A0A182G6Y5 A0A1B6IMK7 Q17HV7 A0A1B0GAB6 A0A1B6CB84 B4JV34 A0A0L7RJF8 A0A154PI07 A0A310S9B8 A0A0A9ZGA3 B0W5X4
PDB
5NVA
E-value=3.06817e-18,
Score=227
Ontologies
GO
Topology
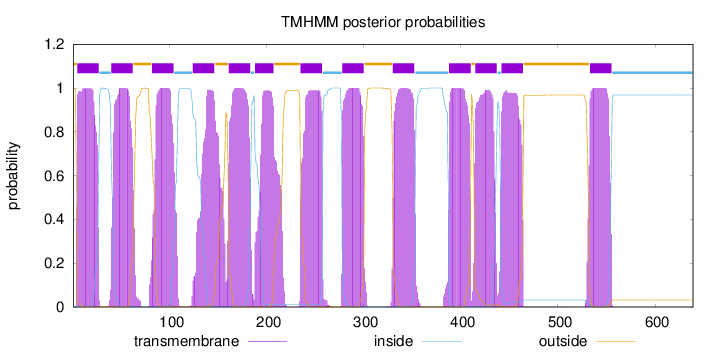
Length:
639
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
287.65831
Exp number, first 60 AAs:
41.25182
Total prob of N-in:
0.00185
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 39
TMhelix
40 - 62
outside
63 - 81
TMhelix
82 - 104
inside
105 - 123
TMhelix
124 - 146
outside
147 - 160
TMhelix
161 - 183
inside
184 - 187
TMhelix
188 - 207
outside
208 - 234
TMhelix
235 - 257
inside
258 - 277
TMhelix
278 - 300
outside
301 - 329
TMhelix
330 - 352
inside
353 - 387
TMhelix
388 - 410
outside
411 - 414
TMhelix
415 - 437
inside
438 - 441
TMhelix
442 - 464
outside
465 - 532
TMhelix
533 - 555
inside
556 - 639
Population Genetic Test Statistics
Pi
311.205764
Theta
188.021129
Tajima's D
1.992742
CLR
0.211184
CSRT
0.87995600219989
Interpretation
Uncertain
