Gene
KWMTBOMO15734
Pre Gene Modal
BGIBMGA002322
Annotation
PREDICTED:_dedicator_of_cytokinesis_protein_1_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.114
Sequence
CDS
ATGATGCAAACAGAATACAACCACAGTGTTGCCGGAGGAGAGAAAGAGAACTACTTGAAGGAGCTAGAGAGCGAGCTGATCGACAAGCTAGACGTGCTGGTGGAGCGGGGGTGCGGCGACCTGCAGTGGCGGGGGCGGTTCGTGACGTCATGCGGCGCGCGGTGCGACGCGGCGGCGGGCGCGCTGCGGGCGTGCGGGCTGGCGCTGGTGGCGGCGGCGGCGCGCCAGCTCGACACGCTGCTGCAGTACCGGGCGGCCGCGCTGCGCTCCCCGACGCCGCACCGCATGCACCTCACCACCACCGTGCTGCACTTCTACCAGGAGATCGAGCGCCCGCACATGTACATACGGTACGTCCACAAGCTGGCCAGCATGTACCGCGCGGCCCAGCAGTGGACCGAGGCTGGGCTCACGCTGCGGCTGCACGCGAAGCTGTTGTCCTGGAGCCACGACCCGCTCCCGCCCAGACTGCGCCATCCCACGCTCAACCACGACGACCACACGACACATAGAGAGCTAAAGGAACATCTCTATTTTGAAATAGCTGAGCTCTTGAAACGCGGCGGTCTCTGGGAGCTGGCGGTCGATATAGTCAAAGAGCTCGTATCGGTTTACGAAGAGGAAGCTCTCGGCTACGGGCCGCTGGCCGAACTGCACAGTCAGCTGGCGGAGTTCTACGCTTCGATGCTGCAGTCCCGGAGGTCGCATCCGGGATACTTCAGGGTCATATATCACGGGAAGGGATTCCCCGAGATATTAAGGAAACCATTGGGCTGCCTCAGCACGATACCGTCGCAGGGCTTCATATACCGCGGCAACGAGTACGAGCAACTCCAGGAGTTCATCGAGAGGATGCTGGACGAGTGGCCCGACGCGGAAGTGTTGCCGCGCCTGGACCCGCCCGGCGAGGATATCACGGAATCCGAAGGACAATATCTTCAAATAAATGCTGTAGATCCGATAATGAGCGATAAGCTCAAACGTTTGTCGGGCAAACCGGTCGCCGAACAGATCCTGCAGTACTACAAGCACAACGACGTCGACAAGTTCTATTTCTCGCGACCCTTCGACCGGCCTGAGGAGTCGTTGAGCGACTCGACTGAAGATTTGAATAAGAATGAGTTCGCGAGACGTTGGCTCGAAAGGACTGAACTAGAGATAAGCGACAAATTGCCCGGCATCCTGCGTTGGTTCCCGGTGGTCAAGCAACGCACGTACTGGGTCTCGCCCATCGAGGTCGCGGTCGAGTCGTTGCGCCAAACGAATAGGACCCTTAAGTCTGTAGCGTTTGAGGCCAGGAACCCCGACACACCCCTCAATCCGCTCACAATGCGATTATTGGGTATCCTCGACTCGGCCGTGCAAGGTGGCATAGTGAACTACGAGAACTCGTTCCTGACCCCGGCCTATGAGAGCCGACACCCGGAGCACGCGGCCCTGCTCGTCGAGCTCAAAGATTTGATCGCCGACCAAATACCGCTGCTCAAGTTCGGCTTAGATGTTCACGCCAGCCGAGCTCCGCCCGCGCAAGAAGAGATGCACGCGCATCTCATGGAACGATTTCGAACCGTACAGAAGCACGTGCACCAGAGATACGGGAGGAAGACTTGCGACTTTGAAACGGAATCGCAAGAAGTTCAACTTCGGATAACTCTGAGAAACAGCCTGCACACGGACACCACGAGTACAGTCACCAACAATCGTATGTCCGACATATCAACCGGAAACGACGTTTCGTCTAAGAGTCCCGGTAGATTCACCAGTTTCAACAGCGCTATAAACCAGTTCGCCAGGAATAGCAGTGGCGGCGTTGCAAACTATCTTGGCACCCTACAGAATCCACCGAAGAAACGTAGTAAAGCACGAGCCGGAAGAAAGAGCGAAGTTGGAACTCAATCGAGCGGCTCCCAAAGCCACTTCTACACTTCGCACGCGACGGTCAATGGCAACGACGCGCGGTCTCTATCCCACAATAGCGGCGTGTGCAACACCCCTAACAGCGTGACGTCATTGAACGAGACCCCGATGATGTCGACGCTACGCGAACTGCGGCAAGAGCTGGTCACCGAGCGACCGCTCCGTGCCGAGGTCGAACGCGAGAAGAGACTGTCCCAACGGGCAAGTCTTTTGACCACTTCTTCCGCACCGCCTTCCAACAGAGACTCGATTGGCACGACCGATTCGAACCAGGACGAAGAGCTGCCACCGCCTTTGCCAATGAAATCCCACCGAAGCATGGAATTGGACAATGACAGTAACAATAACGCCGATCCGACACCATTGCCGGTCCGCCACAACTACGACACGGTTCTTGCGCCGCGAGGATCATTCCTGTACAACTCGATCGTGCACAGAAGGAACACGAATGTAGAGAAGGCGCCGACGCCCCCACCTAAGAAACGTAATGCTCAATGA
Protein
MMQTEYNHSVAGGEKENYLKELESELIDKLDVLVERGCGDLQWRGRFVTSCGARCDAAAGALRACGLALVAAAARQLDTLLQYRAAALRSPTPHRMHLTTTVLHFYQEIERPHMYIRYVHKLASMYRAAQQWTEAGLTLRLHAKLLSWSHDPLPPRLRHPTLNHDDHTTHRELKEHLYFEIAELLKRGGLWELAVDIVKELVSVYEEEALGYGPLAELHSQLAEFYASMLQSRRSHPGYFRVIYHGKGFPEILRKPLGCLSTIPSQGFIYRGNEYEQLQEFIERMLDEWPDAEVLPRLDPPGEDITESEGQYLQINAVDPIMSDKLKRLSGKPVAEQILQYYKHNDVDKFYFSRPFDRPEESLSDSTEDLNKNEFARRWLERTELEISDKLPGILRWFPVVKQRTYWVSPIEVAVESLRQTNRTLKSVAFEARNPDTPLNPLTMRLLGILDSAVQGGIVNYENSFLTPAYESRHPEHAALLVELKDLIADQIPLLKFGLDVHASRAPPAQEEMHAHLMERFRTVQKHVHQRYGRKTCDFETESQEVQLRITLRNSLHTDTTSTVTNNRMSDISTGNDVSSKSPGRFTSFNSAINQFARNSSGGVANYLGTLQNPPKKRSKARAGRKSEVGTQSSGSQSHFYTSHATVNGNDARSLSHNSGVCNTPNSVTSLNETPMMSTLRELRQELVTERPLRAEVEREKRLSQRASLLTTSSAPPSNRDSIGTTDSNQDEELPPPLPMKSHRSMELDNDSNNNADPTPLPVRHNYDTVLAPRGSFLYNSIVHRRNTNVEKAPTPPPKKRNAQ
Summary
Similarity
Belongs to the DOCK family.
Uniprot
H9IYI8
A0A2W1BQC7
A0A2A4K836
A0A2A4K7R2
A0A194QIF2
A0A2J7QWC9
+ More
A0A2J7QWA8 A0A1B6JPF2 A0A2P8Z663 D6X2E2 A0A0N0BK65 A0A310SSU2 A0A195D8Q5 F4X0K6 A0A195D0K2 A0A195F8L0 A0A026WHY8 A0A151XGA9 A0A2S2QES1 A0A1A9Y2K6 E2A054 A0A1A9ZUP3 A0A158ND15 A0A1B0BKX7 A0A195B8U8 A0A1B0FP07 A0A1A9VGF6 A0A1A9WS47 A0A0C9Q9Z8 U4U860 N6TCK0 A0A067RRZ8 B3P8D6 A0A154PH24 A0A0L0CDY5 Q8MQW7 B4N833 A0A0Q9WTI9 A0A0L7RJF5 D0IQF0 A0A1I8PH01 B4M077 B4KDA8 A0A1I8PGS0 A0A0Q9XBS2 A0A0Q9WHR4 A0A1L8DMR3 B4R254 A0A0B4K7Q6 A0A0Q9X8V4 V5NZW8 Q9VCH4 J9K7Q7 A0A1I8NGF6 T1PJX8 T1P9N6 A0A0M4EKB3 A0A034V0Q4 A0A1Y1JWH7 A0A2H8THR5 B4HFZ1 A0A034V4Z8 A0A1J1J3F0 W8AN40 A0A1J1IZ24 A0A3B0JLE7 A0A1J1IZ83 A0A3B0JQ55 A0A3B0JL59 Q299C0 I5ANT7 E0VJW3 A0A034V3W7 B4JTA3 A0A034V5M4 A0A0K8V2S2 A0A0K8WB46 A0A1W4ULM6 A0A1W4U8N1 A0A2S2PFW6 B3LY44 A0A0P9A1P7 B4PNY3 A0A0R1E7L3 W8AJF3 A0A023F393
A0A2J7QWA8 A0A1B6JPF2 A0A2P8Z663 D6X2E2 A0A0N0BK65 A0A310SSU2 A0A195D8Q5 F4X0K6 A0A195D0K2 A0A195F8L0 A0A026WHY8 A0A151XGA9 A0A2S2QES1 A0A1A9Y2K6 E2A054 A0A1A9ZUP3 A0A158ND15 A0A1B0BKX7 A0A195B8U8 A0A1B0FP07 A0A1A9VGF6 A0A1A9WS47 A0A0C9Q9Z8 U4U860 N6TCK0 A0A067RRZ8 B3P8D6 A0A154PH24 A0A0L0CDY5 Q8MQW7 B4N833 A0A0Q9WTI9 A0A0L7RJF5 D0IQF0 A0A1I8PH01 B4M077 B4KDA8 A0A1I8PGS0 A0A0Q9XBS2 A0A0Q9WHR4 A0A1L8DMR3 B4R254 A0A0B4K7Q6 A0A0Q9X8V4 V5NZW8 Q9VCH4 J9K7Q7 A0A1I8NGF6 T1PJX8 T1P9N6 A0A0M4EKB3 A0A034V0Q4 A0A1Y1JWH7 A0A2H8THR5 B4HFZ1 A0A034V4Z8 A0A1J1J3F0 W8AN40 A0A1J1IZ24 A0A3B0JLE7 A0A1J1IZ83 A0A3B0JQ55 A0A3B0JL59 Q299C0 I5ANT7 E0VJW3 A0A034V3W7 B4JTA3 A0A034V5M4 A0A0K8V2S2 A0A0K8WB46 A0A1W4ULM6 A0A1W4U8N1 A0A2S2PFW6 B3LY44 A0A0P9A1P7 B4PNY3 A0A0R1E7L3 W8AJF3 A0A023F393
Pubmed
19121390
28756777
26354079
29403074
18362917
19820115
+ More
21719571 24508170 20798317 21347285 23537049 24845553 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9245788 26109357 26109356 25315136 25348373 28004739 24495485 15632085 20566863 17550304 25474469
21719571 24508170 20798317 21347285 23537049 24845553 17994087 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9245788 26109357 26109356 25315136 25348373 28004739 24495485 15632085 20566863 17550304 25474469
EMBL
BABH01020713
BABH01020714
BABH01020715
KZ150018
PZC74940.1
NWSH01000072
+ More
PCG79953.1 PCG79954.1 KQ458761 KPJ05174.1 NEVH01009765 PNF32874.1 PNF32873.1 GECU01006570 JAT01137.1 PYGN01000176 PSN51994.1 KQ971371 EFA10254.2 KQ435706 KOX80154.1 KQ759821 OAD62690.1 KQ981135 KYN09246.1 GL888498 EGI59998.1 KQ977012 KYN06402.1 KQ981744 KYN36389.1 KK107213 EZA55286.1 KQ982173 KYQ59426.1 GGMS01006499 MBY75702.1 GL435551 EFN73187.1 ADTU01012179 ADTU01012180 ADTU01012181 JXJN01016120 JXJN01016121 KQ976558 KYM80665.1 CCAG010008697 CCAG010008698 GBYB01000044 JAG69811.1 KB632204 ERL90099.1 APGK01043404 KB741014 ENN75463.1 KK852498 KDR22579.1 CH954182 EDV53960.1 KQ434902 KZC11159.1 JRES01000501 KNC30623.1 AY122254 AAM52766.1 CH964232 EDW81284.1 KRF99420.1 KQ414581 KOC70964.1 BT100188 ACY40022.1 CH940650 EDW68327.1 CH933806 EDW14890.2 KRG01267.1 KRF83778.1 GFDF01006454 JAV07630.1 CM000364 EDX14111.1 AE014297 AFH06585.1 KRG01266.1 BT150411 AHA95715.1 AF007805 AAB69648.1 AAF56191.2 ABLF02022160 KA649102 AFP63731.1 KA644648 AFP59277.1 CP012526 ALC47596.1 GAKP01022111 JAC36841.1 GEZM01099147 GEZM01099145 GEZM01099143 JAV53463.1 GFXV01000993 MBW12798.1 CH480815 EDW43384.1 GAKP01022112 JAC36840.1 CVRI01000064 CRL05401.1 GAMC01020652 JAB85903.1 CRL05399.1 OUUW01000007 SPP83055.1 CRL05400.1 SPP83053.1 SPP83054.1 CM000070 EAL27783.2 EIM52622.1 AAZO01002923 DS235231 EEB13669.1 GAKP01022110 JAC36842.1 CH916373 EDV94993.1 GAKP01022109 JAC36843.1 GDHF01019444 JAI32870.1 GDHF01018421 GDHF01004022 JAI33893.1 JAI48292.1 GGMR01015718 MBY28337.1 CH902617 EDV43948.2 KPU80557.1 CM000160 EDW98193.2 KRK04147.1 GAMC01020653 JAB85902.1 GBBI01002984 JAC15728.1
PCG79953.1 PCG79954.1 KQ458761 KPJ05174.1 NEVH01009765 PNF32874.1 PNF32873.1 GECU01006570 JAT01137.1 PYGN01000176 PSN51994.1 KQ971371 EFA10254.2 KQ435706 KOX80154.1 KQ759821 OAD62690.1 KQ981135 KYN09246.1 GL888498 EGI59998.1 KQ977012 KYN06402.1 KQ981744 KYN36389.1 KK107213 EZA55286.1 KQ982173 KYQ59426.1 GGMS01006499 MBY75702.1 GL435551 EFN73187.1 ADTU01012179 ADTU01012180 ADTU01012181 JXJN01016120 JXJN01016121 KQ976558 KYM80665.1 CCAG010008697 CCAG010008698 GBYB01000044 JAG69811.1 KB632204 ERL90099.1 APGK01043404 KB741014 ENN75463.1 KK852498 KDR22579.1 CH954182 EDV53960.1 KQ434902 KZC11159.1 JRES01000501 KNC30623.1 AY122254 AAM52766.1 CH964232 EDW81284.1 KRF99420.1 KQ414581 KOC70964.1 BT100188 ACY40022.1 CH940650 EDW68327.1 CH933806 EDW14890.2 KRG01267.1 KRF83778.1 GFDF01006454 JAV07630.1 CM000364 EDX14111.1 AE014297 AFH06585.1 KRG01266.1 BT150411 AHA95715.1 AF007805 AAB69648.1 AAF56191.2 ABLF02022160 KA649102 AFP63731.1 KA644648 AFP59277.1 CP012526 ALC47596.1 GAKP01022111 JAC36841.1 GEZM01099147 GEZM01099145 GEZM01099143 JAV53463.1 GFXV01000993 MBW12798.1 CH480815 EDW43384.1 GAKP01022112 JAC36840.1 CVRI01000064 CRL05401.1 GAMC01020652 JAB85903.1 CRL05399.1 OUUW01000007 SPP83055.1 CRL05400.1 SPP83053.1 SPP83054.1 CM000070 EAL27783.2 EIM52622.1 AAZO01002923 DS235231 EEB13669.1 GAKP01022110 JAC36842.1 CH916373 EDV94993.1 GAKP01022109 JAC36843.1 GDHF01019444 JAI32870.1 GDHF01018421 GDHF01004022 JAI33893.1 JAI48292.1 GGMR01015718 MBY28337.1 CH902617 EDV43948.2 KPU80557.1 CM000160 EDW98193.2 KRK04147.1 GAMC01020653 JAB85902.1 GBBI01002984 JAC15728.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000235965
UP000245037
UP000007266
+ More
UP000053105 UP000078492 UP000007755 UP000078542 UP000078541 UP000053097 UP000075809 UP000092443 UP000000311 UP000092445 UP000005205 UP000092460 UP000078540 UP000092444 UP000078200 UP000091820 UP000030742 UP000019118 UP000027135 UP000008711 UP000076502 UP000037069 UP000007798 UP000053825 UP000095300 UP000008792 UP000009192 UP000000304 UP000000803 UP000007819 UP000095301 UP000092553 UP000001292 UP000183832 UP000268350 UP000001819 UP000009046 UP000001070 UP000192221 UP000007801 UP000002282
UP000053105 UP000078492 UP000007755 UP000078542 UP000078541 UP000053097 UP000075809 UP000092443 UP000000311 UP000092445 UP000005205 UP000092460 UP000078540 UP000092444 UP000078200 UP000091820 UP000030742 UP000019118 UP000027135 UP000008711 UP000076502 UP000037069 UP000007798 UP000053825 UP000095300 UP000008792 UP000009192 UP000000304 UP000000803 UP000007819 UP000095301 UP000092553 UP000001292 UP000183832 UP000268350 UP000001819 UP000009046 UP000001070 UP000192221 UP000007801 UP000002282
Interpro
Gene 3D
ProteinModelPortal
H9IYI8
A0A2W1BQC7
A0A2A4K836
A0A2A4K7R2
A0A194QIF2
A0A2J7QWC9
+ More
A0A2J7QWA8 A0A1B6JPF2 A0A2P8Z663 D6X2E2 A0A0N0BK65 A0A310SSU2 A0A195D8Q5 F4X0K6 A0A195D0K2 A0A195F8L0 A0A026WHY8 A0A151XGA9 A0A2S2QES1 A0A1A9Y2K6 E2A054 A0A1A9ZUP3 A0A158ND15 A0A1B0BKX7 A0A195B8U8 A0A1B0FP07 A0A1A9VGF6 A0A1A9WS47 A0A0C9Q9Z8 U4U860 N6TCK0 A0A067RRZ8 B3P8D6 A0A154PH24 A0A0L0CDY5 Q8MQW7 B4N833 A0A0Q9WTI9 A0A0L7RJF5 D0IQF0 A0A1I8PH01 B4M077 B4KDA8 A0A1I8PGS0 A0A0Q9XBS2 A0A0Q9WHR4 A0A1L8DMR3 B4R254 A0A0B4K7Q6 A0A0Q9X8V4 V5NZW8 Q9VCH4 J9K7Q7 A0A1I8NGF6 T1PJX8 T1P9N6 A0A0M4EKB3 A0A034V0Q4 A0A1Y1JWH7 A0A2H8THR5 B4HFZ1 A0A034V4Z8 A0A1J1J3F0 W8AN40 A0A1J1IZ24 A0A3B0JLE7 A0A1J1IZ83 A0A3B0JQ55 A0A3B0JL59 Q299C0 I5ANT7 E0VJW3 A0A034V3W7 B4JTA3 A0A034V5M4 A0A0K8V2S2 A0A0K8WB46 A0A1W4ULM6 A0A1W4U8N1 A0A2S2PFW6 B3LY44 A0A0P9A1P7 B4PNY3 A0A0R1E7L3 W8AJF3 A0A023F393
A0A2J7QWA8 A0A1B6JPF2 A0A2P8Z663 D6X2E2 A0A0N0BK65 A0A310SSU2 A0A195D8Q5 F4X0K6 A0A195D0K2 A0A195F8L0 A0A026WHY8 A0A151XGA9 A0A2S2QES1 A0A1A9Y2K6 E2A054 A0A1A9ZUP3 A0A158ND15 A0A1B0BKX7 A0A195B8U8 A0A1B0FP07 A0A1A9VGF6 A0A1A9WS47 A0A0C9Q9Z8 U4U860 N6TCK0 A0A067RRZ8 B3P8D6 A0A154PH24 A0A0L0CDY5 Q8MQW7 B4N833 A0A0Q9WTI9 A0A0L7RJF5 D0IQF0 A0A1I8PH01 B4M077 B4KDA8 A0A1I8PGS0 A0A0Q9XBS2 A0A0Q9WHR4 A0A1L8DMR3 B4R254 A0A0B4K7Q6 A0A0Q9X8V4 V5NZW8 Q9VCH4 J9K7Q7 A0A1I8NGF6 T1PJX8 T1P9N6 A0A0M4EKB3 A0A034V0Q4 A0A1Y1JWH7 A0A2H8THR5 B4HFZ1 A0A034V4Z8 A0A1J1J3F0 W8AN40 A0A1J1IZ24 A0A3B0JLE7 A0A1J1IZ83 A0A3B0JQ55 A0A3B0JL59 Q299C0 I5ANT7 E0VJW3 A0A034V3W7 B4JTA3 A0A034V5M4 A0A0K8V2S2 A0A0K8WB46 A0A1W4ULM6 A0A1W4U8N1 A0A2S2PFW6 B3LY44 A0A0P9A1P7 B4PNY3 A0A0R1E7L3 W8AJF3 A0A023F393
PDB
2YIN
E-value=1.24075e-80,
Score=766
Ontologies
KEGG
GO
PANTHER
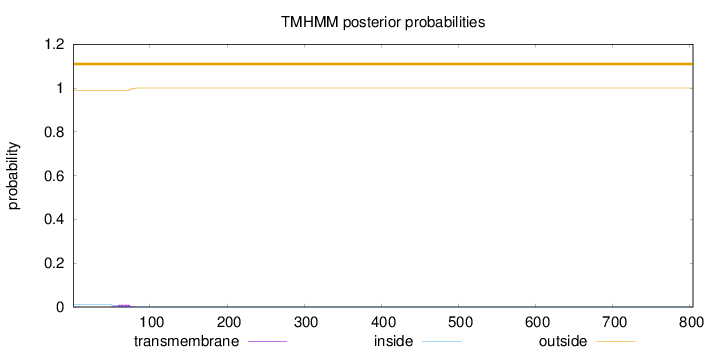
Topology
Length:
804
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19918
Exp number, first 60 AAs:
0.04548
Total prob of N-in:
0.01008
outside
1 - 804
Population Genetic Test Statistics
Pi
279.636995
Theta
165.396622
Tajima's D
2.319991
CLR
0.37176
CSRT
0.928703564821759
Interpretation
Uncertain
