Gene
KWMTBOMO15733
Pre Gene Modal
BGIBMGA002321
Annotation
PREDICTED:_dedicator_of_cytokinesis_protein_1_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.558 Nuclear Reliability : 1.436
Sequence
CDS
ATGACTGTTTGGCGAAAATTAGAAAATGAAGAATGCTATGCCGTTGCAATATACAATTTCACAAGCAAATCCCAGTCGCATCTCCCTTTGGAAGTGGGCGAGCTCATTCATCTCGAAGGTGAAACGGAAGAGTGGTATTGCGGTCAGAGTCTCCGGAGGGACCTCAAAGGCGCCTTCCCAAAAAGTTATGTCGTCAAGAGGGATTGCGTTGTTGACAGATGCAGCGAAAGTGTAGTGGCTTCAGCTGGCGGCTATGGCGTCGTTCACGATATCGCGGTGACATTGCGAGACTGGTTAGAACAATGGAAGAACTTGTATATTAATAATGATGAACGTTTTAAATTTATGGAAGTAAGTATGCGTGCGTTATTAGAGCTAAGAGCTCAGGCCGCCAGCGGAGCGCTTCCTCAAGACCAGCTGCGAAGAGTCGCCAGGACATCTGTATTCACTATCGACAGGGGAAATAGGACGCTAGATATGGAGTTGGCGGTGAGGAATTCAAACGGTCAACTCGTCGATCCGCTGAAGATGTCGACGTACCGTTTGAACGCATTACACGACGAGGCCAACACTCGTATCGATAAAAACATGGAAACGACGCCGGTAACACCGAACGTTACTCCGGCCGGAGTGAGGACGTTCACGGTTTGCGTCCGAGTGCACAACTTCGTGTGTCGAGTGGCGGACGTGGCCGAGCTGTCGCTGTCCTTGCACGACGCGGCCGGGAACAAGTTGACTGAACATCTCCTGATAAGATGGCCCACGCCAAATTTCGCGACCACCAATGTCGTCTTCACGGATTTCGGCAGTGACTTCATGAAGAACGAGAAAGTGTATTTAGTGTGTCACGTCGTGAGAATAGGGTCGATGGACGCTCAAAATGTTGATCACAGGCGCAGTTCAGTGGCCCAGCCGGTGTCGATGAGCACGTCGGCCGGCGCGATGCGGAGGCCGTTCGGCGTGGCCTGCGCCGACATCAAGCGCCACCTCACGTCTGATAACCCTGACAAGCACATACAAGTGCCTTTTTACGCATACGAAAAAGAGAATTTAGATGCTCTGCTGAAGAAGGTGATGGCCAACCGGGAGTTGAAGGACGCCAAGCACTCGCAAGGACTCTGGGTCTCCATACATCTAGTTGAAGGTGATTTAAAACAGATCCGCGAAGAGAATCCGCACATAGTGGTGAACACGGTGGTGGCACGGAAGATGGGCTTCCCCGAAGTGATACTGCCGGGGGACGCGCGCAACGACCTCTACGTGACCCTCTGCTCCGGCTCGTTCTCCAAGGGTGGCGGGAAGTCTTCGGAGAGGAACATCGAGCTGGTGGCCAGGGTTGTTGATCAACACGGGAGAACCATACCGGGTGTGATTTCGATTGGAGCTGGCGTACCGTTAGCTAACGAGTACACTTCGCTTGTGTACTACCATGACGACAGGCCGCGATGGCAGGAAGTGTTTAAGGTGTGTCTGAACATTGAGGAGTTCAAAGACGCTCACCTGGTGTTCCTGTGCAGACACCGGAGCTCGAACGAAGCCAAGGACCGGGCGGAGAGGCATTTCGCGTTAGCCTTCCTCAGGCTCATGCAGCGCGAAGGGACCACGACGCCCGACACCAACCACAACCTGTGCGTTTACAAAATTGACAACAAAAGGTCGAACCCCGCCGAGGTGGAGAAAGAAGCGGCCGCGGTCTGCCTGGGGCTGCCTTCGAAACGGGACGAGCTCCCGGCCGGCGCCGAGCGGGGCCTGACCCGCGGCGTGCTGTCCCTCATACACCGGGACACGCTCACCGTCGCCACCAAGCTGTGCTCCACTAAACTGACGCAGAAAGAGGAAATCCTCGGTGTTCTGAAATGGAGCACGCATCGGACGCCTGGGACATTGCGGCCGGCCTTGACCCAGCTGCTCCACGTCCCGAGCGAAGAGCTGGTCAAGTTCCTCCAGGACATACTGGACACGTTGTTCAGCATACTGACGCAGGTCGAAGAAGACCAAGAATATTATTCGGACAAGAGCTACGGCGTTCTAGTGTTTGAATGCCTGCTGAGGGTCATATCGCTGGTCGCCGATCACAAGTACCAGCATTTCCAGCCAGTTCTGCACGTTTACATAGACGAGAGCTTCTGCGACGCTTTGGCTTACGAGTCCCTGATAACGGTGATGGTGTGGGTGATCCGGACCGCGAACAGCAGCGAGGCGGCCGTCAAGCGCCTGCTGGTCTGCATGAAGTGCGTCGAGAGTCTGCTGCGGCTGGTGGTGCGCTCAAGGCAACTCCGCAGCGCTCTGGGCGACAGCGCTGCTGACGACGGCTTCAAGGCGCTATTCGATACGCTTCTAGAAGCGCTAGTGTGGCTGATGCGGTGCGGCGAGCACGTGCTCACGTGTCAGGGATCCGCGCTGAAGTACTTCCCGCACGCGGTGCCGCACATCATCAAAGTGTACCCGGACACAGAGCTCTGCGCATACATAATCCGTGCATTGGATGCGTTGCCCCTTTGTCGTCTCGGCACTCAAAGGATGCTGGCGTTACTAGAGTTGGTCCGTGGACCACTGTGCGCCGCGCCGGCTGCCAGAGCTTTGCTGCTGCCGCATCTCGCAGCCACACTTCGAGCTTTGTTGAGGGATAAAGCCGAGTGGTACCGCGAGACGGCGCTCAAGTTGCGATTGGTTGGCAAGGCAGCGAGACTCCTAGGCGAGGACACTGCGAAACTACACGATCCAACGCATCACCAGCAAATCGTGGAACTCTGCGTGGAAACCTTAGGCGAGGTGTTGACGCTACTCGCTCGCGATGACGTGGGGCCCGTCGAGAATGATCGAGCGGAATTAGCTAGAAGACTACTGCCCACAGTGCTGAAAACGGCCACCGTGATGATCCAAGAGAGGGGGACCGAGGAACATCCTAGTTCGTCCAATGAGAACATTGTTCGACGAATAGTTTCGGTACTGGTCGACATGGTGCGACAAATGTCCGAAGATCAGTATTCTATGGTGGTGAAAGCGCTTGAAGTGGAAAATGCCGGCGGATCGGGCTCCCTAGTCGACGACGCGCTGGGCCTGATAGTGATGTTGGTTCAGCGACCGGTGTTCAGACCGCACTGGGCGGATATGCTACATCTACAGCATTACGTGATGCTTCATGCTCTGAGGCTGCTGTCAACGACCATTCAAGACCGCCTCAACACGGCCGAAGACACGGAGTCACTGGCCAAAGTCCGCGCACAATGCAAGGCCTGGTTCAGTGCCGGGGCGGCCCTGGGCACGTCGGCGCCTCTCCAGCTCGAGACCTTGACCGCAGCGAGGAAACAACGCGTCAAGCAACTGTACGGGGACATAAGAAGGGGCTTAGCGCAATGCCTCAGCGACATGTGGTACAGCTTGGATGCCGCCAAGATAACACTAGACAAATAA
Protein
MTVWRKLENEECYAVAIYNFTSKSQSHLPLEVGELIHLEGETEEWYCGQSLRRDLKGAFPKSYVVKRDCVVDRCSESVVASAGGYGVVHDIAVTLRDWLEQWKNLYINNDERFKFMEVSMRALLELRAQAASGALPQDQLRRVARTSVFTIDRGNRTLDMELAVRNSNGQLVDPLKMSTYRLNALHDEANTRIDKNMETTPVTPNVTPAGVRTFTVCVRVHNFVCRVADVAELSLSLHDAAGNKLTEHLLIRWPTPNFATTNVVFTDFGSDFMKNEKVYLVCHVVRIGSMDAQNVDHRRSSVAQPVSMSTSAGAMRRPFGVACADIKRHLTSDNPDKHIQVPFYAYEKENLDALLKKVMANRELKDAKHSQGLWVSIHLVEGDLKQIREENPHIVVNTVVARKMGFPEVILPGDARNDLYVTLCSGSFSKGGGKSSERNIELVARVVDQHGRTIPGVISIGAGVPLANEYTSLVYYHDDRPRWQEVFKVCLNIEEFKDAHLVFLCRHRSSNEAKDRAERHFALAFLRLMQREGTTTPDTNHNLCVYKIDNKRSNPAEVEKEAAAVCLGLPSKRDELPAGAERGLTRGVLSLIHRDTLTVATKLCSTKLTQKEEILGVLKWSTHRTPGTLRPALTQLLHVPSEELVKFLQDILDTLFSILTQVEEDQEYYSDKSYGVLVFECLLRVISLVADHKYQHFQPVLHVYIDESFCDALAYESLITVMVWVIRTANSSEAAVKRLLVCMKCVESLLRLVVRSRQLRSALGDSAADDGFKALFDTLLEALVWLMRCGEHVLTCQGSALKYFPHAVPHIIKVYPDTELCAYIIRALDALPLCRLGTQRMLALLELVRGPLCAAPAARALLLPHLAATLRALLRDKAEWYRETALKLRLVGKAARLLGEDTAKLHDPTHHQQIVELCVETLGEVLTLLARDDVGPVENDRAELARRLLPTVLKTATVMIQERGTEEHPSSSNENIVRRIVSVLVDMVRQMSEDQYSMVVKALEVENAGGSGSLVDDALGLIVMLVQRPVFRPHWADMLHLQHYVMLHALRLLSTTIQDRLNTAEDTESLAKVRAQCKAWFSAGAALGTSAPLQLETLTAARKQRVKQLYGDIRRGLAQCLSDMWYSLDAAKITLDK
Summary
Similarity
Belongs to the DOCK family.
Uniprot
A0A2W1BQC7
A0A194QIF2
A0A0L7RJF5
A0A1Y1JYZ4
A0A2A3ETT3
A0A154PH24
+ More
A0A310SSU2 A0A2P8Z663 A0A067RRZ8 A0A3L8DHR7 A0A088AGC7 A0A026WHY8 F4X0K6 A0A158ND15 A0A195B8U8 A0A195D0K2 A0A1Y1JVX0 A0A1Y1K028 A0A1Y1JWH7 E2A054 D6X2E2 A0A139WCG6 A0A195F8L0 A0A1B6BZP6 A0A2J7QWB1 E2B6U6 A0A1B6DG37 A0A2J7QWB4 A0A195D8Q5 J9K7Q7 A0A131YPY9 A0A224YKX2 L7M6G9 A0A131Y023 A0A131XLZ7 A0A1L1YP07 E0VJW3 U4U860 N6TCK0 A0A1L1YP19 A0A0J7L6V7 A0A2M3Z0X4 A0A2M4B880 A0A2R7WCA5 A0A2M4B878 A0A0T6B5S9 A0A2M4CQE8 A0A182FR68 B7P9N2 A0A0A9XU40 A0A1J1IZ24 A0A2M4CQH7 A0A0A9ZEW5 W5JGN7 A0A2M4A2L0 A0A2M4CY53 T1HNF8 A0A0P5HVL2 A0A1L8DMR3 A0A0P6HLY5 E9G3B8 A0A2M4B892 A0A0P4Y2D5 A0A0P5CPW1 A0A0P5ZY24 A0A1J1IZ83 A0A0P5T4Z2 A0A164UU79 A0A0A9Z542 A0A0P5E1U1 A0A0N8ABP7 A0A0A9Z7E6 A0A0P5NK94 A0A1V9XFG6 A0A023F393 A0A0K8W6W1 W8AJF3 G1KGL2 A0A1L8FJ39 A0A2J8JPH4 A0A1L8FEC5 A0A2D0SG33 A0A2D0SER6 A0A0K8WB46 A0A210R280 A0A1I8PGS8 T1P9N6 W5LA00 A0A1V4KI42 A0A1E1X8N9 F7B387 A0A0D9QWR5 A0A151P4J5 A0A218UT94
A0A310SSU2 A0A2P8Z663 A0A067RRZ8 A0A3L8DHR7 A0A088AGC7 A0A026WHY8 F4X0K6 A0A158ND15 A0A195B8U8 A0A195D0K2 A0A1Y1JVX0 A0A1Y1K028 A0A1Y1JWH7 E2A054 D6X2E2 A0A139WCG6 A0A195F8L0 A0A1B6BZP6 A0A2J7QWB1 E2B6U6 A0A1B6DG37 A0A2J7QWB4 A0A195D8Q5 J9K7Q7 A0A131YPY9 A0A224YKX2 L7M6G9 A0A131Y023 A0A131XLZ7 A0A1L1YP07 E0VJW3 U4U860 N6TCK0 A0A1L1YP19 A0A0J7L6V7 A0A2M3Z0X4 A0A2M4B880 A0A2R7WCA5 A0A2M4B878 A0A0T6B5S9 A0A2M4CQE8 A0A182FR68 B7P9N2 A0A0A9XU40 A0A1J1IZ24 A0A2M4CQH7 A0A0A9ZEW5 W5JGN7 A0A2M4A2L0 A0A2M4CY53 T1HNF8 A0A0P5HVL2 A0A1L8DMR3 A0A0P6HLY5 E9G3B8 A0A2M4B892 A0A0P4Y2D5 A0A0P5CPW1 A0A0P5ZY24 A0A1J1IZ83 A0A0P5T4Z2 A0A164UU79 A0A0A9Z542 A0A0P5E1U1 A0A0N8ABP7 A0A0A9Z7E6 A0A0P5NK94 A0A1V9XFG6 A0A023F393 A0A0K8W6W1 W8AJF3 G1KGL2 A0A1L8FJ39 A0A2J8JPH4 A0A1L8FEC5 A0A2D0SG33 A0A2D0SER6 A0A0K8WB46 A0A210R280 A0A1I8PGS8 T1P9N6 W5LA00 A0A1V4KI42 A0A1E1X8N9 F7B387 A0A0D9QWR5 A0A151P4J5 A0A218UT94
Pubmed
EMBL
KZ150018
PZC74940.1
KQ458761
KPJ05174.1
KQ414581
KOC70964.1
+ More
GEZM01099152 JAV53458.1 KZ288191 PBC34461.1 KQ434902 KZC11159.1 KQ759821 OAD62690.1 PYGN01000176 PSN51994.1 KK852498 KDR22579.1 QOIP01000007 RLU19995.1 KK107213 EZA55286.1 GL888498 EGI59998.1 ADTU01012179 ADTU01012180 ADTU01012181 KQ976558 KYM80665.1 KQ977012 KYN06402.1 GEZM01099151 GEZM01099148 GEZM01099144 JAV53459.1 GEZM01099150 JAV53460.1 GEZM01099147 GEZM01099145 GEZM01099143 JAV53463.1 GL435551 EFN73187.1 KQ971371 EFA10254.2 KYB25596.1 KQ981744 KYN36389.1 GEDC01030838 JAS06460.1 NEVH01009765 PNF32870.1 GL446009 EFN88587.1 GEDC01012723 JAS24575.1 PNF32871.1 KQ981135 KYN09246.1 ABLF02022160 GEDV01007929 JAP80628.1 GFPF01007130 MAA18276.1 GACK01005409 JAA59625.1 GEFM01004460 GEGO01002417 JAP71336.1 JAR92987.1 GEFH01001865 JAP66716.1 KP100156 AKO62846.1 AAZO01002923 DS235231 EEB13669.1 KB632204 ERL90099.1 APGK01043404 KB741014 ENN75463.1 AKO62845.1 LBMM01000472 KMQ98259.1 GGFM01001436 MBW22187.1 GGFJ01000113 MBW49254.1 KK854560 PTY16871.1 GGFJ01000112 MBW49253.1 LJIG01009602 KRT82752.1 GGFL01003384 MBW67562.1 ABJB010131106 ABJB010191271 ABJB010349701 ABJB010747103 ABJB010816281 ABJB010870676 ABJB010919045 ABJB010939443 ABJB011013157 ABJB011087767 DS665503 EEC03304.1 GBHO01019397 JAG24207.1 CVRI01000064 CRL05399.1 GGFL01003414 MBW67592.1 GBHO01003214 JAG40390.1 ADMH02001587 ETN61959.1 GGFK01001712 MBW35033.1 GGFL01005620 MBW69798.1 ACPB03007651 GDIQ01224526 JAK27199.1 GFDF01006454 JAV07630.1 GDIQ01018373 JAN76364.1 GL732531 EFX86015.1 GGFJ01000128 MBW49269.1 GDIP01237307 JAI86094.1 GDIP01167160 JAJ56242.1 GDIP01037433 JAM66282.1 CRL05400.1 GDIP01133225 JAL70489.1 LRGB01001574 KZS11673.1 GBHO01003217 JAG40387.1 GDIP01147664 JAJ75738.1 GDIP01163161 JAJ60241.1 GBHO01003215 JAG40389.1 GDIQ01145102 JAL06624.1 MNPL01012059 OQR72305.1 GBBI01002984 JAC15728.1 GDHF01005453 JAI46861.1 GAMC01020653 JAB85902.1 CM004478 OCT71603.1 NBAG03000436 PNI24637.1 CM004479 OCT69949.1 GDHF01018421 GDHF01004022 JAI33893.1 JAI48292.1 NEDP02000770 OWF55042.1 KA644648 AFP59277.1 LSYS01003057 OPJ84061.1 GFAC01003802 JAT95386.1 AAMC01014969 AAMC01014970 AAMC01014971 AAMC01014972 AAMC01014973 AAMC01014974 AAMC01014975 AAMC01014976 AAMC01014977 AAMC01014978 AQIB01042219 AQIB01042220 AQIB01042221 AQIB01042222 AQIB01042223 AQIB01042224 AKHW03001049 KYO43900.1 MUZQ01000141 OWK56965.1
GEZM01099152 JAV53458.1 KZ288191 PBC34461.1 KQ434902 KZC11159.1 KQ759821 OAD62690.1 PYGN01000176 PSN51994.1 KK852498 KDR22579.1 QOIP01000007 RLU19995.1 KK107213 EZA55286.1 GL888498 EGI59998.1 ADTU01012179 ADTU01012180 ADTU01012181 KQ976558 KYM80665.1 KQ977012 KYN06402.1 GEZM01099151 GEZM01099148 GEZM01099144 JAV53459.1 GEZM01099150 JAV53460.1 GEZM01099147 GEZM01099145 GEZM01099143 JAV53463.1 GL435551 EFN73187.1 KQ971371 EFA10254.2 KYB25596.1 KQ981744 KYN36389.1 GEDC01030838 JAS06460.1 NEVH01009765 PNF32870.1 GL446009 EFN88587.1 GEDC01012723 JAS24575.1 PNF32871.1 KQ981135 KYN09246.1 ABLF02022160 GEDV01007929 JAP80628.1 GFPF01007130 MAA18276.1 GACK01005409 JAA59625.1 GEFM01004460 GEGO01002417 JAP71336.1 JAR92987.1 GEFH01001865 JAP66716.1 KP100156 AKO62846.1 AAZO01002923 DS235231 EEB13669.1 KB632204 ERL90099.1 APGK01043404 KB741014 ENN75463.1 AKO62845.1 LBMM01000472 KMQ98259.1 GGFM01001436 MBW22187.1 GGFJ01000113 MBW49254.1 KK854560 PTY16871.1 GGFJ01000112 MBW49253.1 LJIG01009602 KRT82752.1 GGFL01003384 MBW67562.1 ABJB010131106 ABJB010191271 ABJB010349701 ABJB010747103 ABJB010816281 ABJB010870676 ABJB010919045 ABJB010939443 ABJB011013157 ABJB011087767 DS665503 EEC03304.1 GBHO01019397 JAG24207.1 CVRI01000064 CRL05399.1 GGFL01003414 MBW67592.1 GBHO01003214 JAG40390.1 ADMH02001587 ETN61959.1 GGFK01001712 MBW35033.1 GGFL01005620 MBW69798.1 ACPB03007651 GDIQ01224526 JAK27199.1 GFDF01006454 JAV07630.1 GDIQ01018373 JAN76364.1 GL732531 EFX86015.1 GGFJ01000128 MBW49269.1 GDIP01237307 JAI86094.1 GDIP01167160 JAJ56242.1 GDIP01037433 JAM66282.1 CRL05400.1 GDIP01133225 JAL70489.1 LRGB01001574 KZS11673.1 GBHO01003217 JAG40387.1 GDIP01147664 JAJ75738.1 GDIP01163161 JAJ60241.1 GBHO01003215 JAG40389.1 GDIQ01145102 JAL06624.1 MNPL01012059 OQR72305.1 GBBI01002984 JAC15728.1 GDHF01005453 JAI46861.1 GAMC01020653 JAB85902.1 CM004478 OCT71603.1 NBAG03000436 PNI24637.1 CM004479 OCT69949.1 GDHF01018421 GDHF01004022 JAI33893.1 JAI48292.1 NEDP02000770 OWF55042.1 KA644648 AFP59277.1 LSYS01003057 OPJ84061.1 GFAC01003802 JAT95386.1 AAMC01014969 AAMC01014970 AAMC01014971 AAMC01014972 AAMC01014973 AAMC01014974 AAMC01014975 AAMC01014976 AAMC01014977 AAMC01014978 AQIB01042219 AQIB01042220 AQIB01042221 AQIB01042222 AQIB01042223 AQIB01042224 AKHW03001049 KYO43900.1 MUZQ01000141 OWK56965.1
Proteomes
UP000053268
UP000053825
UP000242457
UP000076502
UP000245037
UP000027135
+ More
UP000279307 UP000005203 UP000053097 UP000007755 UP000005205 UP000078540 UP000078542 UP000000311 UP000007266 UP000078541 UP000235965 UP000008237 UP000078492 UP000007819 UP000009046 UP000030742 UP000019118 UP000036403 UP000069272 UP000001555 UP000183832 UP000000673 UP000015103 UP000000305 UP000076858 UP000192247 UP000001646 UP000186698 UP000221080 UP000242188 UP000095300 UP000018467 UP000190648 UP000008143 UP000029965 UP000050525 UP000197619
UP000279307 UP000005203 UP000053097 UP000007755 UP000005205 UP000078540 UP000078542 UP000000311 UP000007266 UP000078541 UP000235965 UP000008237 UP000078492 UP000007819 UP000009046 UP000030742 UP000019118 UP000036403 UP000069272 UP000001555 UP000183832 UP000000673 UP000015103 UP000000305 UP000076858 UP000192247 UP000001646 UP000186698 UP000221080 UP000242188 UP000095300 UP000018467 UP000190648 UP000008143 UP000029965 UP000050525 UP000197619
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BQC7
A0A194QIF2
A0A0L7RJF5
A0A1Y1JYZ4
A0A2A3ETT3
A0A154PH24
+ More
A0A310SSU2 A0A2P8Z663 A0A067RRZ8 A0A3L8DHR7 A0A088AGC7 A0A026WHY8 F4X0K6 A0A158ND15 A0A195B8U8 A0A195D0K2 A0A1Y1JVX0 A0A1Y1K028 A0A1Y1JWH7 E2A054 D6X2E2 A0A139WCG6 A0A195F8L0 A0A1B6BZP6 A0A2J7QWB1 E2B6U6 A0A1B6DG37 A0A2J7QWB4 A0A195D8Q5 J9K7Q7 A0A131YPY9 A0A224YKX2 L7M6G9 A0A131Y023 A0A131XLZ7 A0A1L1YP07 E0VJW3 U4U860 N6TCK0 A0A1L1YP19 A0A0J7L6V7 A0A2M3Z0X4 A0A2M4B880 A0A2R7WCA5 A0A2M4B878 A0A0T6B5S9 A0A2M4CQE8 A0A182FR68 B7P9N2 A0A0A9XU40 A0A1J1IZ24 A0A2M4CQH7 A0A0A9ZEW5 W5JGN7 A0A2M4A2L0 A0A2M4CY53 T1HNF8 A0A0P5HVL2 A0A1L8DMR3 A0A0P6HLY5 E9G3B8 A0A2M4B892 A0A0P4Y2D5 A0A0P5CPW1 A0A0P5ZY24 A0A1J1IZ83 A0A0P5T4Z2 A0A164UU79 A0A0A9Z542 A0A0P5E1U1 A0A0N8ABP7 A0A0A9Z7E6 A0A0P5NK94 A0A1V9XFG6 A0A023F393 A0A0K8W6W1 W8AJF3 G1KGL2 A0A1L8FJ39 A0A2J8JPH4 A0A1L8FEC5 A0A2D0SG33 A0A2D0SER6 A0A0K8WB46 A0A210R280 A0A1I8PGS8 T1P9N6 W5LA00 A0A1V4KI42 A0A1E1X8N9 F7B387 A0A0D9QWR5 A0A151P4J5 A0A218UT94
A0A310SSU2 A0A2P8Z663 A0A067RRZ8 A0A3L8DHR7 A0A088AGC7 A0A026WHY8 F4X0K6 A0A158ND15 A0A195B8U8 A0A195D0K2 A0A1Y1JVX0 A0A1Y1K028 A0A1Y1JWH7 E2A054 D6X2E2 A0A139WCG6 A0A195F8L0 A0A1B6BZP6 A0A2J7QWB1 E2B6U6 A0A1B6DG37 A0A2J7QWB4 A0A195D8Q5 J9K7Q7 A0A131YPY9 A0A224YKX2 L7M6G9 A0A131Y023 A0A131XLZ7 A0A1L1YP07 E0VJW3 U4U860 N6TCK0 A0A1L1YP19 A0A0J7L6V7 A0A2M3Z0X4 A0A2M4B880 A0A2R7WCA5 A0A2M4B878 A0A0T6B5S9 A0A2M4CQE8 A0A182FR68 B7P9N2 A0A0A9XU40 A0A1J1IZ24 A0A2M4CQH7 A0A0A9ZEW5 W5JGN7 A0A2M4A2L0 A0A2M4CY53 T1HNF8 A0A0P5HVL2 A0A1L8DMR3 A0A0P6HLY5 E9G3B8 A0A2M4B892 A0A0P4Y2D5 A0A0P5CPW1 A0A0P5ZY24 A0A1J1IZ83 A0A0P5T4Z2 A0A164UU79 A0A0A9Z542 A0A0P5E1U1 A0A0N8ABP7 A0A0A9Z7E6 A0A0P5NK94 A0A1V9XFG6 A0A023F393 A0A0K8W6W1 W8AJF3 G1KGL2 A0A1L8FJ39 A0A2J8JPH4 A0A1L8FEC5 A0A2D0SG33 A0A2D0SER6 A0A0K8WB46 A0A210R280 A0A1I8PGS8 T1P9N6 W5LA00 A0A1V4KI42 A0A1E1X8N9 F7B387 A0A0D9QWR5 A0A151P4J5 A0A218UT94
PDB
3L4C
E-value=3.37319e-34,
Score=367
Ontologies
KEGG
GO
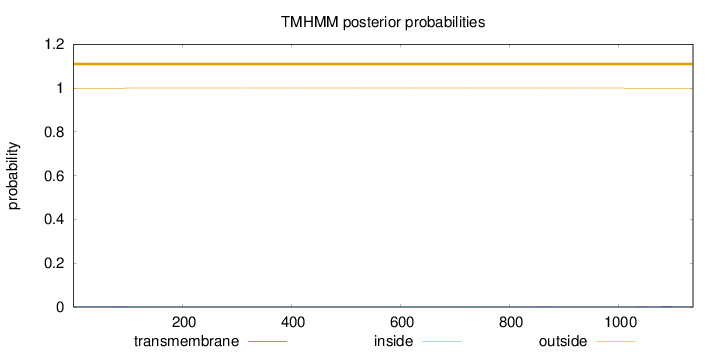
Topology
Length:
1137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0680199999999999
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.00085
outside
1 - 1137
Population Genetic Test Statistics
Pi
233.036879
Theta
175.749083
Tajima's D
1.271527
CLR
0.354521
CSRT
0.731113444327784
Interpretation
Uncertain
