Gene
KWMTBOMO15730
Pre Gene Modal
BGIBMGA002257
Annotation
PREDICTED:_RNA-directed_DNA_polymerase_from_mobile_element_jockey-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.704
Sequence
CDS
ATGCAATTTCTCAGTGATCCCTGCGCTAAGCTAGAATCTTTGTCAGCTGAGGTGCCAAGTACAGCTCGTAAAGGAAAGGGTAAAGAAAATCGAGTGTTAAACGACATTATTGCCATCTTTAAAACAGAACCTGATGCGTTGCCAGTGTTTGTGGCCCGAGATCTCAACAAGTTACCACCAATCACCTTCGATCACTTGGATGTTTCCAAACTACTAAAAGATCTCGTGCTGGTCCAAGCTGAAATTAAGGCCATTAAATCGTCGTACGTGACTCAGGATGAAATGCAAGACTTGAGGACAAAGTGTGACATGATCTCAATGTCTTCGACAAACATAAATACAAAGAGAGGTGCTTTTTGTAATAAAATTGACATGGAGTTTACGAGTCAAGAAAGTAATTCTGGTTGCGAGGAGCAAGTATTAATAAAAAACTTAAGCTCAAATTCTACAAATACCGAAAATCTAATAGAAGGTAGTGGCATAAAACTTCGACGGTATGAATTGAACGGTAGTGACAACAACTTATCGCACCACAACGTAGCGAGTAAAGGTAACGGGAACGACGAGTCATTGCTATCGTCCGTCGATCAGCTGAGTGAAAGGATTATAGTGCCGTCACTGATCGCACCGAGTCAATCGTATGCTAAAGTTGTAGCAGTACAAAATAACGATGGATTCACTTTGGTACAATCGAAGAAAAAACGTAACAATAAGTTTAAAGGCAGTGTAGGTAGTGTAGTAATAGAATCAAACGAGAAGTTTAGGTCCGCTGATCGAAAAACGCCTATGTTCATAACAAACGTAAACAAGGCAACATTAGAATCAGATATAAATGACTACATCTACAAGAAAACAGGAGAGAATGTGACCTTAGAACGTATCAACATCAAACGGCAATGTGAACACAATGCGTACAAGTTTATTGTGTCCCAAAGAAAGCTGCATATGTTTATGGATGATAAGTTATGGCCTGAGGGAATTATTTTCCGACGATTTGTACATTACAGACCGAAACGTAATAACGAGACTGTGTACATGCCAACAGACATTGCGGCAAATCTTACGGATTTTACGGATATCCTGAGTGTGGTGAGTGCCATTATTGAGAGCTGTCATATTGACCATGTTTTTATGTTGGGTGATTTTAATGCACATCCTTCCGAACAATTTTATGATGAGTTATCTGTATATTGTGATGAACAAAAATGGATATGTATAGATACTAAATTATTAGGAATACGATCTAACACGTATACTTTCTTAAGCGAGGCTCATATGTCAAAGAGGTGGCTTGATCATTGCGTGGTTACCGAAGCTGCTGTGAGCTTGGTTGAGAATGTATACGTGTTGTATGACGTGTTATGGTCCGATCACTATCCTTTGATAGTTGAATGTAGGTTAGATAAAATAACTGCAAAAAGACTTGATGTTTGTTTAAGTAAATCCAATAACGTTATTTGGGGTGAGAGAAGTCCTGATCAAATAGAATCTTATCAGAAAGAATGTAATAAAAGATTAAGATTAATTGACTTTCCTCATGAACTCAGTAAATGCTGTGATGGTGTGTGTACTGAAGTAAATCACATAGTAGTTATAGATAAATTGTATACAGATATTGTAAATGCCCTAAGTGCAGCTGCATTGGACGGTCGGGAGGGCGCTGTAAAACGTGCTAGAAGAGTGGTAGTGGGCTGGAATAAGCATGTTGCCGAGGCTCACAGGGAAGCTAGGCAAAAGTTTTTGTTGTGGGTAGGGTACGGTAAACCTACTTCTGGGTATGTGTATAATGATATGTGTGAATCTAGAAAAGTTTTCAAGTCTAGACTAAAGTGGTGCCAAGATCATGAGGACCAAATAAAAATGGATATTCTAGCCTCACACCATTCTAAAAGTGATTTTCGTGGCTTTTGGAAACACACAAATAAATTAAAGAATAGGCCGACAATACCTGTGAGCGTCGGTGGCGTGAGTGAACCAGAACAAATTGCCAACATGTTCAAGGAACGCTTCTTTGTAAAATCTACTTTAGGACTCGCTGGGTCGGTGCTTGATATTGAGCACGGCAGGGAAATTGGAGTGGCATTTACGTCTAAGGATGTTAAAAAGACAATTAGGTCAATGTCACGTGGTAAGTCTCCGGGACATGACGGGCTCAGCATTGAGCACCTGCGTTATGCGGGCGTGCACATCTCTAGGGTTCTCTCAATGCTGTATAACATTTGTGTGGGTCATAGTTATTTGCCAAGGGAAATGATACGAACAGTAGTTGTACCAATAGCTAAAAATAAGACTGGAGACCTTGCAGATAGCAGTAACTATAGGCCTATCTCACTAGCTACTATTATATCCAAAGTGTTGGACAGCATGCTTAACTTTAAACTAAATAAATATGTAAATCTTCATGATAACCAATTCGGATTCCGTTCAGGTTTGTCGACAGAGAGTGCAATCTTAAGTCTTAAGCATGCTGTCAAATACTACACGCGGCGAGGAACACCTGTTTATGCCTGCTTTCTTGATCTGTCCAAGGCGTTTGATTTGGTTTCCTATGACCTCTTGTGGAAAAAGTTGGAAAATATTAAACTACCCACGGAAATAAATAACATTCTAAAGTTTTGGTATGGGAACCAGATCAACAATGTGCGATGGGCTGGGGCTTTGTCTCTTCCATATAGGTTGGAGTGCGGGGTGAGACAAGGCGGGTTGAGTTCGCCTACGCTCTTCAACCTGTATATAAACGAGCTGATCGGCGAGCTCAGTAGGACCAGGATTGGTTGTTTCATAGACGGGGTGTGCGTAAACAACATCAGCTACGCAGACGATATGGTGCTGCTGAGCGCGTCTATCTGTGGCCTAAGGCAACTTGTTTCTTTGTGTGAGGGATACGCCAAATCACACGGTTTGGTGTACAACTGTAAAAAGTCGGAGATCATGGTCTTTGAGACCAGGGGTGGGACGCATGATAAAATACCTCCCGTAGTACTCAACGGAACGCCATTGACAAGAGTTTATAAATTTAAATATCTCGGCCATGTGTTGACCCCTGATCTCAAAGATGATGAAGATATTGAACGAGAGCGGAGAGCGTTGTCGGTTAGAGCAAACATGATAGCTCGCAGGTTTGCACGGTGTTCACTTAAAGTCAAGGTCACCCTCTTTAGGGCCTACTGTACTAACTTTTACACGTGCAGCTTGTGGGCTGGATATACTCAAAGATCGTACAATGCTCTCCGTGTTCAATACAATAACGCGTTTAGGGTGCTGGTGGTGCTGCCCCGCTTTTGTAGCGCATCAGGGATGTTTGCAGACGCGCACATAGATTGTTTTTACGCTACCATGCGTAAGAGGTGTGCATCTCTGGTAAACAGAGTGAGGGCCAGCTCCAACAGTATCCTGAGCATGATCGCAAGCAGGCTGGACTGTATATACATGGGTCGCTGTGGTGCCATCTCCCATGGGATGTTACCACAGTAA
Protein
MQFLSDPCAKLESLSAEVPSTARKGKGKENRVLNDIIAIFKTEPDALPVFVARDLNKLPPITFDHLDVSKLLKDLVLVQAEIKAIKSSYVTQDEMQDLRTKCDMISMSSTNINTKRGAFCNKIDMEFTSQESNSGCEEQVLIKNLSSNSTNTENLIEGSGIKLRRYELNGSDNNLSHHNVASKGNGNDESLLSSVDQLSERIIVPSLIAPSQSYAKVVAVQNNDGFTLVQSKKKRNNKFKGSVGSVVIESNEKFRSADRKTPMFITNVNKATLESDINDYIYKKTGENVTLERINIKRQCEHNAYKFIVSQRKLHMFMDDKLWPEGIIFRRFVHYRPKRNNETVYMPTDIAANLTDFTDILSVVSAIIESCHIDHVFMLGDFNAHPSEQFYDELSVYCDEQKWICIDTKLLGIRSNTYTFLSEAHMSKRWLDHCVVTEAAVSLVENVYVLYDVLWSDHYPLIVECRLDKITAKRLDVCLSKSNNVIWGERSPDQIESYQKECNKRLRLIDFPHELSKCCDGVCTEVNHIVVIDKLYTDIVNALSAAALDGREGAVKRARRVVVGWNKHVAEAHREARQKFLLWVGYGKPTSGYVYNDMCESRKVFKSRLKWCQDHEDQIKMDILASHHSKSDFRGFWKHTNKLKNRPTIPVSVGGVSEPEQIANMFKERFFVKSTLGLAGSVLDIEHGREIGVAFTSKDVKKTIRSMSRGKSPGHDGLSIEHLRYAGVHISRVLSMLYNICVGHSYLPREMIRTVVVPIAKNKTGDLADSSNYRPISLATIISKVLDSMLNFKLNKYVNLHDNQFGFRSGLSTESAILSLKHAVKYYTRRGTPVYACFLDLSKAFDLVSYDLLWKKLENIKLPTEINNILKFWYGNQINNVRWAGALSLPYRLECGVRQGGLSSPTLFNLYINELIGELSRTRIGCFIDGVCVNNISYADDMVLLSASICGLRQLVSLCEGYAKSHGLVYNCKKSEIMVFETRGGTHDKIPPVVLNGTPLTRVYKFKYLGHVLTPDLKDDEDIERERRALSVRANMIARRFARCSLKVKVTLFRAYCTNFYTCSLWAGYTQRSYNALRVQYNNAFRVLVVLPRFCSASGMFADAHIDCFYATMRKRCASLVNRVRASSNSILSMIASRLDCIYMGRCGAISHGMLPQ
Summary
Uniprot
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
6AR3
E-value=3.739e-05,
Score=117
Ontologies
GO
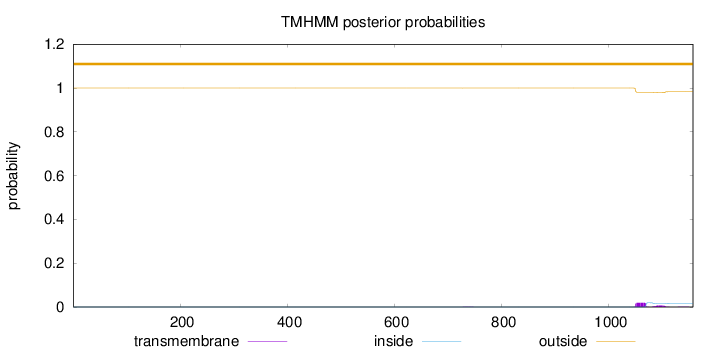
Topology
Length:
1157
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.51651
Exp number, first 60 AAs:
0.00066
Total prob of N-in:
0.00005
outside
1 - 1157
Population Genetic Test Statistics
Pi
235.200618
Theta
206.634236
Tajima's D
1.387694
CLR
137.694773
CSRT
0.765811709414529
Interpretation
Uncertain
