Pre Gene Modal
BGIBMGA002318
Annotation
high_temperature_requirement_protein_A2_[Bombyx_mori]
Full name
Serine protease HTRA2, mitochondrial
Alternative Name
High temperature requirement protein A2
Omi stress-regulated endoprotease
Omi stress-regulated endoprotease
Location in the cell
Mitochondrial Reliability : 1.531 PlasmaMembrane Reliability : 1.011
Sequence
CDS
ATGAATTTAAGGCTTTTCCGTATTTTTAAAACAAATACTATATCCAAAATAAACTACAGATGTACTAGTCGCGTTGTGCCAATACTAAATAACTACAAAAGTAACAGTGATGACAATCAAAGCTCTTACTTTAGATGCATTTTAGGAGCCACTATAGGGTTTATAGGTTATTTTAGTTTACGTGAAAAGGTAACAGCTGCTACAGTTGTAAATGATTTGAAAGGACGAAGGGAGAAATATAATTTTATAGCTGACGTTGTGGCAGTTTCGGCGGCGTCTGTTGTGTATATAGAAATAGTAGACGGACGAAGAATAGATGCGTTTACCGGTAAAAAATTAAAGATATCCAATGGCTCGGGATTCATCATTAAGGAAGATGGGCTGATATTAACTAACGCTCATGTTGTAGTCAATAAGCCTAATGCAATCGTCAAAGTTAGATTAACTGATGGTTCAACACACGAGGCCCTCATAGAACACTATGACCTACAATCAGATCTAGCAACACTTCGGATACCAGTCAAAGGACTACCAACAATGAAACTTGGTACTTCCGCTGATCTCAAGCCTGGCGAGTGGGTGGTGGCTATAGGAAGTCCCCTTGATTTGAGCAACACAGTCACAGCTGGTGTGGTGAGCTCTACTCAGAGAGCTGGAAGCGAACTTGGCTTACAAGACAGAAATATAGTATACATACAAACAGATGCTCCGATTACATTCGGTAACAGCGGGGGACCTCTTGTCAATCTCGACGGCGAAGCTATAGGCATTAACAGCATGAAAGTCACCTACGGAATATCGTTTGCCATACCCATAGATTATGTAAAGGAATTCTTGGCTAAACATAAAACCAAGTCTCCTCAAGTCTCGAAGCGTTACCTTGGCATCACCATGCTCTCATTAACGCCTTCCATACTCATGGAGTTGAAGATGAGAAATCCCGAGATGCCAACCGATATTCAACACGGCATCCTTGTATGGAAGGTCATCATTGGATCGCCGGCTTTCAACGGGGGTCTACAACCAGGCGACATTGTAGTCAAAATAAACGGAAAACCAGTTCACAATACAACCGATATATACAACATTCTAGAAAGTACGACGGGGAGTTTGAAGATTGACGCCGTCAGAGGGCGCCAGCAGATAAACCTGACCATAGTACCAGAGTTACACTGA
Protein
MNLRLFRIFKTNTISKINYRCTSRVVPILNNYKSNSDDNQSSYFRCILGATIGFIGYFSLREKVTAATVVNDLKGRREKYNFIADVVAVSAASVVYIEIVDGRRIDAFTGKKLKISNGSGFIIKEDGLILTNAHVVVNKPNAIVKVRLTDGSTHEALIEHYDLQSDLATLRIPVKGLPTMKLGTSADLKPGEWVVAIGSPLDLSNTVTAGVVSSTQRAGSELGLQDRNIVYIQTDAPITFGNSGGPLVNLDGEAIGINSMKVTYGISFAIPIDYVKEFLAKHKTKSPQVSKRYLGITMLSLTPSILMELKMRNPEMPTDIQHGILVWKVIIGSPAFNGGLQPGDIVVKINGKPVHNTTDIYNILESTTGSLKIDAVRGRQQINLTIVPELH
Summary
Description
Serine protease that shows proteolytic activity against a non-specific substrate beta-casein. Promotes or induces cell death either by direct binding to and inhibition of BIRC proteins (also called inhibitor of apoptosis proteins, IAPs), leading to an increase in caspase activity, or by a BIRC inhibition-independent, caspase-independent and serine protease activity-dependent mechanism. Can antagonize antiapoptotic activity of th by directly inducing the degradation of th (By similarity).
Serine protease that shows proteolytic activity against a non-specific substrate beta-casein. Promotes or induces cell death either by direct binding to and inhibition of BIRC proteins (also called inhibitor of apoptosis proteins, IAPs), leading to an increase in caspase activity, or by a BIRC inhibition-independent, caspase-independent and serine protease activity-dependent mechanism. Can antagonize antiapoptotic activity of th by directly inducing the degradation of th.
Serine protease that shows proteolytic activity against a non-specific substrate beta-casein. Promotes or induces cell death either by direct binding to and inhibition of BIRC proteins (also called inhibitor of apoptosis proteins, IAPs), leading to an increase in caspase activity, or by a BIRC inhibition-independent, caspase-independent and serine protease activity-dependent mechanism. Can antagonize antiapoptotic activity of th by directly inducing the degradation of th.
Catalytic Activity
Cleavage of non-polar aliphatic amino-acids at the P1 position, with a preference for Val, Ile and Met. At the P2 and P3 positions, Arg is selected most strongly with a secondary preference for other hydrophilic residues.
Subunit
Interacts with th/DIAP1 (via BIR 2 domain).
Similarity
Belongs to the peptidase S1C family.
Keywords
Apoptosis
Complete proteome
Hydrolase
Membrane
Mitochondrion
Protease
Reference proteome
Serine protease
Transit peptide
Transmembrane
Transmembrane helix
Zymogen
Direct protein sequencing
Ubl conjugation
Feature
chain Serine protease HTRA2, mitochondrial
Uniprot
E9JEH9
A0A2A4JQZ8
A0A2W1BPY2
A0A2A4JQ82
A0A194QNW3
A0A2H1WSF0
+ More
A0A0N1IGF8 B4G316 Q297U2 A0A0M5J9A9 D6WGF2 A0A1W4USN6 A0A0B4KG52 Q9VFJ3 B4HEM8 B4K835 B4QZU6 B4LY58 B3LVG7 B4PST0 B3P3J9 Q176F8 A0A023ET05 A0A3B0JDW7 Q176F7 A0A1S4FDA2 A0A336L1Z8 A0A1Y1KS05 A0A0K8VBF4 W8C2G7 A0A0A1WGS0 A0A1Q3F965 A0A182H2K1 B4N937 A0A336M9M5 A0A034W811 A0A0K8TK95 A0A0K8VAR6 A0A1Q3FBA9 B4JTT7 A0A1A9UEQ4 A0A1A9X9Z1 A0A1B0BH89 A0A1B0G0K2 A0A1A9ZMD8 A0A084VLM0 A0A1A9WNT6 A0A1I8PU85 A0A182KS70 Q7QF03 A0A182XPG7 A0A182VJQ7 A0A1I8M2X9 A0A182UBC6 A0A182IAG4 A0A0L0CA47 A0A1J1HLP9 A0A2J7RFQ5 K7IR33 A0A232EY33 A0A182M124 A0A154PIY5 A0A1B0D3U5 A0A1L8DTK2 A0A1B0CSJ1 A0A182IKI5 A0A182N0B5 A0A182QEE8 A0A182XY18 A0A1V1FKJ0 A0A182RQ72 A0A182T6P8 N6T759 A0A182W597 A0A067RIC9 A0A182K9C3 A0A2M4BRC5 A0A2M4BR44 A0A2M4BR51 A0A2M3ZAZ8 W5JAK0 E2AUP0 A0A2M4ALR3 A0A2M4ANZ4 E2BYN0 A0A151IDG5 A0A182FJ30 A0A151WEF3 T1HZX5 A0A224XFH9 E9IAJ5 A0A195EE27 A0A195FFQ2 A0A182PR27 A0A195BEW1 A0A158NUD5 T1IQJ2 B0WE06 A0A068F678 F4X495
A0A0N1IGF8 B4G316 Q297U2 A0A0M5J9A9 D6WGF2 A0A1W4USN6 A0A0B4KG52 Q9VFJ3 B4HEM8 B4K835 B4QZU6 B4LY58 B3LVG7 B4PST0 B3P3J9 Q176F8 A0A023ET05 A0A3B0JDW7 Q176F7 A0A1S4FDA2 A0A336L1Z8 A0A1Y1KS05 A0A0K8VBF4 W8C2G7 A0A0A1WGS0 A0A1Q3F965 A0A182H2K1 B4N937 A0A336M9M5 A0A034W811 A0A0K8TK95 A0A0K8VAR6 A0A1Q3FBA9 B4JTT7 A0A1A9UEQ4 A0A1A9X9Z1 A0A1B0BH89 A0A1B0G0K2 A0A1A9ZMD8 A0A084VLM0 A0A1A9WNT6 A0A1I8PU85 A0A182KS70 Q7QF03 A0A182XPG7 A0A182VJQ7 A0A1I8M2X9 A0A182UBC6 A0A182IAG4 A0A0L0CA47 A0A1J1HLP9 A0A2J7RFQ5 K7IR33 A0A232EY33 A0A182M124 A0A154PIY5 A0A1B0D3U5 A0A1L8DTK2 A0A1B0CSJ1 A0A182IKI5 A0A182N0B5 A0A182QEE8 A0A182XY18 A0A1V1FKJ0 A0A182RQ72 A0A182T6P8 N6T759 A0A182W597 A0A067RIC9 A0A182K9C3 A0A2M4BRC5 A0A2M4BR44 A0A2M4BR51 A0A2M3ZAZ8 W5JAK0 E2AUP0 A0A2M4ALR3 A0A2M4ANZ4 E2BYN0 A0A151IDG5 A0A182FJ30 A0A151WEF3 T1HZX5 A0A224XFH9 E9IAJ5 A0A195EE27 A0A195FFQ2 A0A182PR27 A0A195BEW1 A0A158NUD5 T1IQJ2 B0WE06 A0A068F678 F4X495
EC Number
3.4.21.108
Pubmed
19121390
21040523
28756777
26354079
17994087
15632085
+ More
18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17397804 17557079 18259196 12537569 17510324 24945155 28004739 24495485 25830018 26483478 25348373 26369729 24438588 20966253 12364791 14747013 17210077 25315136 26108605 20075255 28648823 25244985 28410430 23537049 24845553 20920257 23761445 20798317 21282665 21347285 24952583 21719571
18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17397804 17557079 18259196 12537569 17510324 24945155 28004739 24495485 25830018 26483478 25348373 26369729 24438588 20966253 12364791 14747013 17210077 25315136 26108605 20075255 28648823 25244985 28410430 23537049 24845553 20920257 23761445 20798317 21282665 21347285 24952583 21719571
EMBL
BABH01020677
BABH01020678
BABH01020679
GQ426296
ADM32522.1
NWSH01000788
+ More
PCG74196.1 KZ150018 PZC74936.1 PCG74197.1 KQ458761 KPJ05171.1 ODYU01010704 SOQ55983.1 KQ460820 KPJ12097.1 CH479179 EDW24211.1 CM000070 EAL28113.1 CP012526 ALC46598.1 KQ971321 EFA00185.1 AE014297 AGB95946.1 AB112473 AY075206 AAF55062.1 AAL68074.1 BAE72064.1 CH480815 EDW42185.1 CH933806 EDW15389.1 CM000364 EDX12944.1 CH940650 EDW67946.1 CH902617 EDV41495.1 CM000160 EDW97576.1 CH954181 EDV48887.1 CH477387 EAT42058.1 GAPW01002124 JAC11474.1 OUUW01000005 SPP80275.1 EAT42059.1 UFQS01000843 UFQT01000843 SSX07220.1 SSX27563.1 GEZM01075222 JAV64199.1 GDHF01016040 JAI36274.1 GAMC01003137 JAC03419.1 GBXI01016210 JAC98081.1 GFDL01010963 JAV24082.1 JXUM01105531 JXUM01105532 JXUM01105533 KQ564974 KXJ71478.1 CH964232 EDW81584.1 UFQT01000749 SSX26945.1 GAKP01008496 GAKP01008495 JAC50457.1 GDAI01002876 JAI14727.1 GDHF01016378 JAI35936.1 GFDL01010262 JAV24783.1 CH916374 EDV91516.1 JXJN01014221 CCAG010004854 ATLV01014525 KE524974 KFB38864.1 AAAB01008846 EAA06411.5 APCN01001235 JRES01000789 KNC28319.1 CVRI01000010 CRK88983.1 NEVH01004410 PNF39662.1 NNAY01001695 OXU23199.1 AXCM01000049 KQ434902 KZC11170.1 AJVK01023706 GFDF01004308 JAV09776.1 AJWK01026143 AXCN02001229 FX985289 BAX07302.1 APGK01041367 KB740993 ENN76049.1 KK852458 KDR23547.1 GGFJ01006413 MBW55554.1 GGFJ01006414 MBW55555.1 GGFJ01006415 MBW55556.1 GGFM01004966 MBW25717.1 ADMH02001819 ETN61021.1 GL442872 EFN62839.1 GGFK01008227 MBW41548.1 GGFK01009172 MBW42493.1 GL451499 EFN79184.1 KQ977963 KYM98420.1 KQ983238 KYQ46205.1 ACPB03009692 GFTR01005199 JAW11227.1 GL762022 EFZ22436.1 KQ979039 KYN23361.1 KQ981606 KYN39530.1 KQ976509 KYM82727.1 ADTU01000443 ADTU01000444 AFFK01018331 DS231903 EDS45136.1 KJ512094 AID60317.1 GL888633 EGI58691.1
PCG74196.1 KZ150018 PZC74936.1 PCG74197.1 KQ458761 KPJ05171.1 ODYU01010704 SOQ55983.1 KQ460820 KPJ12097.1 CH479179 EDW24211.1 CM000070 EAL28113.1 CP012526 ALC46598.1 KQ971321 EFA00185.1 AE014297 AGB95946.1 AB112473 AY075206 AAF55062.1 AAL68074.1 BAE72064.1 CH480815 EDW42185.1 CH933806 EDW15389.1 CM000364 EDX12944.1 CH940650 EDW67946.1 CH902617 EDV41495.1 CM000160 EDW97576.1 CH954181 EDV48887.1 CH477387 EAT42058.1 GAPW01002124 JAC11474.1 OUUW01000005 SPP80275.1 EAT42059.1 UFQS01000843 UFQT01000843 SSX07220.1 SSX27563.1 GEZM01075222 JAV64199.1 GDHF01016040 JAI36274.1 GAMC01003137 JAC03419.1 GBXI01016210 JAC98081.1 GFDL01010963 JAV24082.1 JXUM01105531 JXUM01105532 JXUM01105533 KQ564974 KXJ71478.1 CH964232 EDW81584.1 UFQT01000749 SSX26945.1 GAKP01008496 GAKP01008495 JAC50457.1 GDAI01002876 JAI14727.1 GDHF01016378 JAI35936.1 GFDL01010262 JAV24783.1 CH916374 EDV91516.1 JXJN01014221 CCAG010004854 ATLV01014525 KE524974 KFB38864.1 AAAB01008846 EAA06411.5 APCN01001235 JRES01000789 KNC28319.1 CVRI01000010 CRK88983.1 NEVH01004410 PNF39662.1 NNAY01001695 OXU23199.1 AXCM01000049 KQ434902 KZC11170.1 AJVK01023706 GFDF01004308 JAV09776.1 AJWK01026143 AXCN02001229 FX985289 BAX07302.1 APGK01041367 KB740993 ENN76049.1 KK852458 KDR23547.1 GGFJ01006413 MBW55554.1 GGFJ01006414 MBW55555.1 GGFJ01006415 MBW55556.1 GGFM01004966 MBW25717.1 ADMH02001819 ETN61021.1 GL442872 EFN62839.1 GGFK01008227 MBW41548.1 GGFK01009172 MBW42493.1 GL451499 EFN79184.1 KQ977963 KYM98420.1 KQ983238 KYQ46205.1 ACPB03009692 GFTR01005199 JAW11227.1 GL762022 EFZ22436.1 KQ979039 KYN23361.1 KQ981606 KYN39530.1 KQ976509 KYM82727.1 ADTU01000443 ADTU01000444 AFFK01018331 DS231903 EDS45136.1 KJ512094 AID60317.1 GL888633 EGI58691.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000008744
UP000001819
+ More
UP000092553 UP000007266 UP000192221 UP000000803 UP000001292 UP000009192 UP000000304 UP000008792 UP000007801 UP000002282 UP000008711 UP000008820 UP000268350 UP000069940 UP000249989 UP000007798 UP000001070 UP000078200 UP000092443 UP000092460 UP000092444 UP000092445 UP000030765 UP000091820 UP000095300 UP000075882 UP000007062 UP000076407 UP000075903 UP000095301 UP000075902 UP000075840 UP000037069 UP000183832 UP000235965 UP000002358 UP000215335 UP000075883 UP000076502 UP000092462 UP000092461 UP000075880 UP000075884 UP000075886 UP000076408 UP000075900 UP000075901 UP000019118 UP000075920 UP000027135 UP000075881 UP000000673 UP000000311 UP000008237 UP000078542 UP000069272 UP000075809 UP000015103 UP000078492 UP000078541 UP000075885 UP000078540 UP000005205 UP000002320 UP000007755
UP000092553 UP000007266 UP000192221 UP000000803 UP000001292 UP000009192 UP000000304 UP000008792 UP000007801 UP000002282 UP000008711 UP000008820 UP000268350 UP000069940 UP000249989 UP000007798 UP000001070 UP000078200 UP000092443 UP000092460 UP000092444 UP000092445 UP000030765 UP000091820 UP000095300 UP000075882 UP000007062 UP000076407 UP000075903 UP000095301 UP000075902 UP000075840 UP000037069 UP000183832 UP000235965 UP000002358 UP000215335 UP000075883 UP000076502 UP000092462 UP000092461 UP000075880 UP000075884 UP000075886 UP000076408 UP000075900 UP000075901 UP000019118 UP000075920 UP000027135 UP000075881 UP000000673 UP000000311 UP000008237 UP000078542 UP000069272 UP000075809 UP000015103 UP000078492 UP000078541 UP000075885 UP000078540 UP000005205 UP000002320 UP000007755
Interpro
ProteinModelPortal
E9JEH9
A0A2A4JQZ8
A0A2W1BPY2
A0A2A4JQ82
A0A194QNW3
A0A2H1WSF0
+ More
A0A0N1IGF8 B4G316 Q297U2 A0A0M5J9A9 D6WGF2 A0A1W4USN6 A0A0B4KG52 Q9VFJ3 B4HEM8 B4K835 B4QZU6 B4LY58 B3LVG7 B4PST0 B3P3J9 Q176F8 A0A023ET05 A0A3B0JDW7 Q176F7 A0A1S4FDA2 A0A336L1Z8 A0A1Y1KS05 A0A0K8VBF4 W8C2G7 A0A0A1WGS0 A0A1Q3F965 A0A182H2K1 B4N937 A0A336M9M5 A0A034W811 A0A0K8TK95 A0A0K8VAR6 A0A1Q3FBA9 B4JTT7 A0A1A9UEQ4 A0A1A9X9Z1 A0A1B0BH89 A0A1B0G0K2 A0A1A9ZMD8 A0A084VLM0 A0A1A9WNT6 A0A1I8PU85 A0A182KS70 Q7QF03 A0A182XPG7 A0A182VJQ7 A0A1I8M2X9 A0A182UBC6 A0A182IAG4 A0A0L0CA47 A0A1J1HLP9 A0A2J7RFQ5 K7IR33 A0A232EY33 A0A182M124 A0A154PIY5 A0A1B0D3U5 A0A1L8DTK2 A0A1B0CSJ1 A0A182IKI5 A0A182N0B5 A0A182QEE8 A0A182XY18 A0A1V1FKJ0 A0A182RQ72 A0A182T6P8 N6T759 A0A182W597 A0A067RIC9 A0A182K9C3 A0A2M4BRC5 A0A2M4BR44 A0A2M4BR51 A0A2M3ZAZ8 W5JAK0 E2AUP0 A0A2M4ALR3 A0A2M4ANZ4 E2BYN0 A0A151IDG5 A0A182FJ30 A0A151WEF3 T1HZX5 A0A224XFH9 E9IAJ5 A0A195EE27 A0A195FFQ2 A0A182PR27 A0A195BEW1 A0A158NUD5 T1IQJ2 B0WE06 A0A068F678 F4X495
A0A0N1IGF8 B4G316 Q297U2 A0A0M5J9A9 D6WGF2 A0A1W4USN6 A0A0B4KG52 Q9VFJ3 B4HEM8 B4K835 B4QZU6 B4LY58 B3LVG7 B4PST0 B3P3J9 Q176F8 A0A023ET05 A0A3B0JDW7 Q176F7 A0A1S4FDA2 A0A336L1Z8 A0A1Y1KS05 A0A0K8VBF4 W8C2G7 A0A0A1WGS0 A0A1Q3F965 A0A182H2K1 B4N937 A0A336M9M5 A0A034W811 A0A0K8TK95 A0A0K8VAR6 A0A1Q3FBA9 B4JTT7 A0A1A9UEQ4 A0A1A9X9Z1 A0A1B0BH89 A0A1B0G0K2 A0A1A9ZMD8 A0A084VLM0 A0A1A9WNT6 A0A1I8PU85 A0A182KS70 Q7QF03 A0A182XPG7 A0A182VJQ7 A0A1I8M2X9 A0A182UBC6 A0A182IAG4 A0A0L0CA47 A0A1J1HLP9 A0A2J7RFQ5 K7IR33 A0A232EY33 A0A182M124 A0A154PIY5 A0A1B0D3U5 A0A1L8DTK2 A0A1B0CSJ1 A0A182IKI5 A0A182N0B5 A0A182QEE8 A0A182XY18 A0A1V1FKJ0 A0A182RQ72 A0A182T6P8 N6T759 A0A182W597 A0A067RIC9 A0A182K9C3 A0A2M4BRC5 A0A2M4BR44 A0A2M4BR51 A0A2M3ZAZ8 W5JAK0 E2AUP0 A0A2M4ALR3 A0A2M4ANZ4 E2BYN0 A0A151IDG5 A0A182FJ30 A0A151WEF3 T1HZX5 A0A224XFH9 E9IAJ5 A0A195EE27 A0A195FFQ2 A0A182PR27 A0A195BEW1 A0A158NUD5 T1IQJ2 B0WE06 A0A068F678 F4X495
PDB
5M3O
E-value=6.80177e-79,
Score=748
Ontologies
PATHWAY
GO
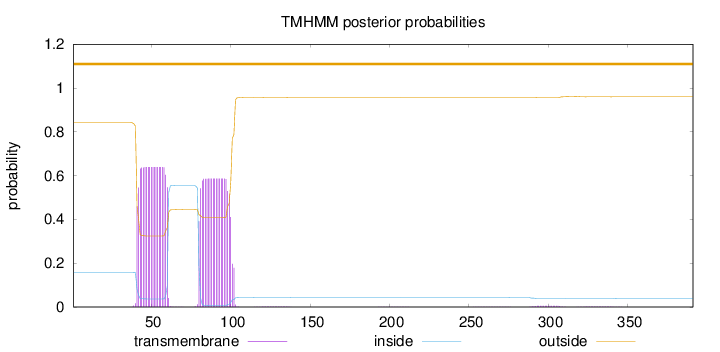
Topology
Subcellular location
Mitochondrion intermembrane space
Predominantly present in the intermembrane space. Released into the cytosol following apoptotic stimuli, such as UV treatment. The extramitochondrial protein does not diffuse throughout the cytosol but stays near the mitochondria. With evidence from 2 publications.
Mitochondrion membrane Predominantly present in the intermembrane space. Released into the cytosol following apoptotic stimuli, such as UV treatment. The extramitochondrial protein does not diffuse throughout the cytosol but stays near the mitochondria. With evidence from 2 publications.
Mitochondrion membrane Predominantly present in the intermembrane space. Released into the cytosol following apoptotic stimuli, such as UV treatment. The extramitochondrial protein does not diffuse throughout the cytosol but stays near the mitochondria. With evidence from 2 publications.
Length:
391
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
24.77581
Exp number, first 60 AAs:
12.39054
Total prob of N-in:
0.15764
POSSIBLE N-term signal
sequence
outside
1 - 391
Population Genetic Test Statistics
Pi
194.166632
Theta
158.875052
Tajima's D
1.213389
CLR
0.080148
CSRT
0.715714214289285
Interpretation
Uncertain
