Pre Gene Modal
BGIBMGA002318
Annotation
PREDICTED:_serine_protease_HTRA2?_mitochondrial-like_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 2.33
Sequence
CDS
ATGTTTTTCGGTGGAGTAGAGGGAGGAGCAACACACTCGAACCTTGTGATATGCGACGAAGCTGGACGGGTTGTTGGGAGAGCTAAAGGTCTCGGCACGAACCATTGGACTCTTGGAATAGACGGATGCGCCAACAGGATCATAAGCATGCTGCACGAGGCCAAAGAAGACGCAGGGATCCCCAAAGATCAAGCCCTAGATTCTTTGGGTTTGACGTTATCAGGCTGTGAACAAGAAAGCAGCAATGCCGAATTAGTGGCCAGGGTCAAAGACTTAGATCCGATGTGTGCAAAAGCGGTGTATGCGGCGTCCGACACGGCTGGGTCCCTGTTCACTGGAGCACCGGACGGAGGCATGGTTCTTATTGCTGGTACCGGTTCGAATGCACTATTACGTACGTCGGATGGAGAACAACACAACTGCGGTGGCTGGGGACATTTGCTCGGCGACGAAGGTGGAGCTTACTGGATAGCGCATAAAGCCGTGAAATCGGTGATAGATGACGTCGATGGTCTCCGTATCTCTCCATATCCAACGCACAACGTATGGGAAGTGATCAGGGAACATTTCGATGCTGACACCAGAGCGGATCTGCTTCCACACGCCTATAAGAACTTTAATAAATCGCAATTTGCGGGTTTAACGGTAAAGCTATCGGCATTGGCATACAAAGGTGACGAGCTATCGAGGCATATCTTCGCCGAAGCTGGTAGTGCGTTAGCGGCTCACGTGGTTGCGCTGGCCAGACGGAGCACCGCCAAAAGGCTGAGAGTCGTCTGCGTTGGATCGGTGTGGAACAGCTGGGACGTGTTGAAACCGGGAATGCTAAATGAACTTAACGCTAAAAAGGTGAAGTGCGAATTGGAACTGGTACGCTTAAAAGTATCGAGCGCAATGGGAGCGGCCTGGTTGGCGGCTAACAAAATAAACTACGACCTACCGAGAGACGACGAGGCCTTCTGTCAAGTATTCCACAAGTACCGACCAGATGCAGTTAATGGTGATGCGAAGGTCAAACACAACGGTCAAGTTAATGGGAATTTAAACGGTGTCGAAAACTGCGGTATAACGTTAAGTCTCTTGTAA
Protein
MFFGGVEGGATHSNLVICDEAGRVVGRAKGLGTNHWTLGIDGCANRIISMLHEAKEDAGIPKDQALDSLGLTLSGCEQESSNAELVARVKDLDPMCAKAVYAASDTAGSLFTGAPDGGMVLIAGTGSNALLRTSDGEQHNCGGWGHLLGDEGGAYWIAHKAVKSVIDDVDGLRISPYPTHNVWEVIREHFDADTRADLLPHAYKNFNKSQFAGLTVKLSALAYKGDELSRHIFAEAGSALAAHVVALARRSTAKRLRVVCVGSVWNSWDVLKPGMLNELNAKKVKCELELVRLKVSSAMGAAWLAANKINYDLPRDDEAFCQVFHKYRPDAVNGDAKVKHNGQVNGNLNGVENCGITLSLL
Summary
Uniprot
A0A2W1BMT5
A0A2A4JQ51
A0A194QJS9
A0A1E1WQC2
S4NXX5
A0A212FB90
+ More
A0A0N1IGF8 A0A1E1W9Q7 B4N7Y5 A0A0K8VRT4 A0A0K8VPD3 B3P0V1 A0A0K8W618 B4PS46 Q9VF86 A0A0L0CRT5 A0A0A1WV85 B4QYJ0 B4HDX4 B3MSU7 A0A1I8N2U8 T1PAP2 A0A1I8PD57 A0A0K8TTK3 A0A034WBN8 A0A1W4VSU9 B4MBK1 A0A1L8EG10 W8BG72 Q299G1 A0A1L8E248 B4JS05 A0A3B0K0E1 A0A0M4F6R8 B4G546 B4KCY1 A0A1I8Q7R0 A0A182Q6L9 A0A182W596 A0A336MD05 A0A182JA65 A0A182H2K0 A0A182IAG5 A0A023ERE5 Q7QF04 A0A023ESK4 A0A2M4AAR5 A0A2M4BSL2 A0A182N0B6 A0A2M4BSL7 A0A2M3ZI14 A0A182MK31 A0A1S4FD93 A0A023EQA5 Q176F9 A0A336N1G9 A0A182VAA2 A0A2J7RFQ6 A0A182FJ31 A0A182PR26 A0A1J1HLP6 W5J988 A0A336MEI2 A0A084VLL9 A0A1Q3FAU4 A0A1A9Y8L6 A0A1B0BRF7 A0A067RI65 A0A1Q3FB23 A0A026W6X4 A0A182XPG6 A0A182TW90 D3TQA1 A0A182K9C2 A0A310SPG5 A0A1B0A2Q0 A0A1Y1L9G4 A0A182XY19 A0A182RQ73 A0A088AMU5 B0WE05 A0A2A3EF77 A0A0M8ZZH5 A0A1A9VLK6 A0A182T5D8 A0A151WMI2 A0A151IZ45 A0A195D497 E9IWS3 K7IMA4 A0A0J7NXF9 A0A195FXD6 A0A336LI78 H0XZM5 H7C3G9 A0A170ZF18
A0A0N1IGF8 A0A1E1W9Q7 B4N7Y5 A0A0K8VRT4 A0A0K8VPD3 B3P0V1 A0A0K8W618 B4PS46 Q9VF86 A0A0L0CRT5 A0A0A1WV85 B4QYJ0 B4HDX4 B3MSU7 A0A1I8N2U8 T1PAP2 A0A1I8PD57 A0A0K8TTK3 A0A034WBN8 A0A1W4VSU9 B4MBK1 A0A1L8EG10 W8BG72 Q299G1 A0A1L8E248 B4JS05 A0A3B0K0E1 A0A0M4F6R8 B4G546 B4KCY1 A0A1I8Q7R0 A0A182Q6L9 A0A182W596 A0A336MD05 A0A182JA65 A0A182H2K0 A0A182IAG5 A0A023ERE5 Q7QF04 A0A023ESK4 A0A2M4AAR5 A0A2M4BSL2 A0A182N0B6 A0A2M4BSL7 A0A2M3ZI14 A0A182MK31 A0A1S4FD93 A0A023EQA5 Q176F9 A0A336N1G9 A0A182VAA2 A0A2J7RFQ6 A0A182FJ31 A0A182PR26 A0A1J1HLP6 W5J988 A0A336MEI2 A0A084VLL9 A0A1Q3FAU4 A0A1A9Y8L6 A0A1B0BRF7 A0A067RI65 A0A1Q3FB23 A0A026W6X4 A0A182XPG6 A0A182TW90 D3TQA1 A0A182K9C2 A0A310SPG5 A0A1B0A2Q0 A0A1Y1L9G4 A0A182XY19 A0A182RQ73 A0A088AMU5 B0WE05 A0A2A3EF77 A0A0M8ZZH5 A0A1A9VLK6 A0A182T5D8 A0A151WMI2 A0A151IZ45 A0A195D497 E9IWS3 K7IMA4 A0A0J7NXF9 A0A195FXD6 A0A336LI78 H0XZM5 H7C3G9 A0A170ZF18
Pubmed
28756777
26354079
23622113
22118469
17994087
17550304
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 25830018 18057021 25315136 26369729 25348373 24495485 15632085 26483478 24945155 12364791 14747013 17210077 17510324 20920257 23761445 24438588 24845553 24508170 20353571 28004739 25244985 21282665 20075255 15815621 18669648 19690332 20068231 21269460 22223895 22814378 23186163 24275569
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 25830018 18057021 25315136 26369729 25348373 24495485 15632085 26483478 24945155 12364791 14747013 17210077 17510324 20920257 23761445 24438588 24845553 24508170 20353571 28004739 25244985 21282665 20075255 15815621 18669648 19690332 20068231 21269460 22223895 22814378 23186163 24275569
EMBL
KZ150018
PZC74935.1
NWSH01000788
PCG74195.1
KQ458761
KPJ05170.1
+ More
GDQN01001845 JAT89209.1 GAIX01011982 JAA80578.1 AGBW02009366 OWR50989.1 KQ460820 KPJ12097.1 GDQN01007314 JAT83740.1 CH964232 EDW81236.1 GDHF01011054 GDHF01010233 JAI41260.1 JAI42081.1 GDHF01025775 GDHF01011580 GDHF01001265 JAI26539.1 JAI40734.1 JAI51049.1 CH954181 EDV48999.1 GDHF01005671 JAI46643.1 CM000160 EDW97466.1 AE014297 AY069061 AAF55174.1 AAL39206.1 JRES01000007 KNC34926.1 GBXI01014542 GBXI01011308 JAC99749.1 JAD02984.1 CM000364 EDX12835.1 CH480815 EDW42066.1 CH902623 EDV30337.1 KPU72804.1 KA645807 AFP60436.1 GDAI01000120 JAI17483.1 GAKP01006838 JAC52114.1 CH940656 EDW58472.1 GFDG01001286 JAV17513.1 GAMC01006256 GAMC01006255 JAC00301.1 CM000070 EAL27742.2 GFDF01001432 JAV12652.1 CH916373 EDV94545.1 OUUW01000013 SPP87775.1 CP012526 ALC47787.1 CH479179 EDW24712.1 CH933806 EDW17003.1 KRG02448.1 AXCN02001229 UFQT01000878 SSX27790.1 JXUM01105531 KQ564974 KXJ71477.1 APCN01001235 APCN01001236 GAPW01001843 JAC11755.1 AAAB01008846 EAA06412.4 GAPW01001842 JAC11756.1 GGFK01004552 MBW37873.1 GGFJ01006935 MBW56076.1 GGFJ01006934 MBW56075.1 GGFM01007456 MBW28207.1 AXCM01000049 GAPW01001841 JAC11757.1 CH477387 EAT42057.1 UFQT01003965 SSX35399.1 NEVH01004410 PNF39663.1 CVRI01000010 CRK88984.1 ADMH02001819 ETN61022.1 SSX27791.1 ATLV01014524 ATLV01014525 KE524974 KFB38863.1 GFDL01010375 JAV24670.1 JXJN01019080 KK852458 KDR23546.1 GFDL01010274 JAV24771.1 KK107390 EZA51376.1 CCAG010021518 EZ423603 ADD19879.1 KQ760549 OAD59903.1 GEZM01062734 JAV69508.1 DS231903 EDS45135.1 KZ288271 PBC29806.1 KQ435812 KOX72719.1 KQ982944 KYQ49056.1 KQ980725 KYN14023.1 KQ976870 KYN07707.1 GL766597 EFZ14978.1 LBMM01000996 KMQ97090.1 KQ981169 KYN45330.1 UFQS01004575 UFQT01004575 SSX16407.1 SSX35719.1 AAQR03102088 AC007881 GEMB01002277 JAS00904.1
GDQN01001845 JAT89209.1 GAIX01011982 JAA80578.1 AGBW02009366 OWR50989.1 KQ460820 KPJ12097.1 GDQN01007314 JAT83740.1 CH964232 EDW81236.1 GDHF01011054 GDHF01010233 JAI41260.1 JAI42081.1 GDHF01025775 GDHF01011580 GDHF01001265 JAI26539.1 JAI40734.1 JAI51049.1 CH954181 EDV48999.1 GDHF01005671 JAI46643.1 CM000160 EDW97466.1 AE014297 AY069061 AAF55174.1 AAL39206.1 JRES01000007 KNC34926.1 GBXI01014542 GBXI01011308 JAC99749.1 JAD02984.1 CM000364 EDX12835.1 CH480815 EDW42066.1 CH902623 EDV30337.1 KPU72804.1 KA645807 AFP60436.1 GDAI01000120 JAI17483.1 GAKP01006838 JAC52114.1 CH940656 EDW58472.1 GFDG01001286 JAV17513.1 GAMC01006256 GAMC01006255 JAC00301.1 CM000070 EAL27742.2 GFDF01001432 JAV12652.1 CH916373 EDV94545.1 OUUW01000013 SPP87775.1 CP012526 ALC47787.1 CH479179 EDW24712.1 CH933806 EDW17003.1 KRG02448.1 AXCN02001229 UFQT01000878 SSX27790.1 JXUM01105531 KQ564974 KXJ71477.1 APCN01001235 APCN01001236 GAPW01001843 JAC11755.1 AAAB01008846 EAA06412.4 GAPW01001842 JAC11756.1 GGFK01004552 MBW37873.1 GGFJ01006935 MBW56076.1 GGFJ01006934 MBW56075.1 GGFM01007456 MBW28207.1 AXCM01000049 GAPW01001841 JAC11757.1 CH477387 EAT42057.1 UFQT01003965 SSX35399.1 NEVH01004410 PNF39663.1 CVRI01000010 CRK88984.1 ADMH02001819 ETN61022.1 SSX27791.1 ATLV01014524 ATLV01014525 KE524974 KFB38863.1 GFDL01010375 JAV24670.1 JXJN01019080 KK852458 KDR23546.1 GFDL01010274 JAV24771.1 KK107390 EZA51376.1 CCAG010021518 EZ423603 ADD19879.1 KQ760549 OAD59903.1 GEZM01062734 JAV69508.1 DS231903 EDS45135.1 KZ288271 PBC29806.1 KQ435812 KOX72719.1 KQ982944 KYQ49056.1 KQ980725 KYN14023.1 KQ976870 KYN07707.1 GL766597 EFZ14978.1 LBMM01000996 KMQ97090.1 KQ981169 KYN45330.1 UFQS01004575 UFQT01004575 SSX16407.1 SSX35719.1 AAQR03102088 AC007881 GEMB01002277 JAS00904.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000007798
UP000008711
+ More
UP000002282 UP000000803 UP000037069 UP000000304 UP000001292 UP000007801 UP000095301 UP000095300 UP000192221 UP000008792 UP000001819 UP000001070 UP000268350 UP000092553 UP000008744 UP000009192 UP000075886 UP000075920 UP000075880 UP000069940 UP000249989 UP000075840 UP000007062 UP000075884 UP000075883 UP000008820 UP000075903 UP000235965 UP000069272 UP000075885 UP000183832 UP000000673 UP000030765 UP000092443 UP000092460 UP000027135 UP000053097 UP000076407 UP000075902 UP000092444 UP000075881 UP000092445 UP000076408 UP000075900 UP000005203 UP000002320 UP000242457 UP000053105 UP000078200 UP000075901 UP000075809 UP000078492 UP000078542 UP000002358 UP000036403 UP000078541 UP000005225 UP000005640
UP000002282 UP000000803 UP000037069 UP000000304 UP000001292 UP000007801 UP000095301 UP000095300 UP000192221 UP000008792 UP000001819 UP000001070 UP000268350 UP000092553 UP000008744 UP000009192 UP000075886 UP000075920 UP000075880 UP000069940 UP000249989 UP000075840 UP000007062 UP000075884 UP000075883 UP000008820 UP000075903 UP000235965 UP000069272 UP000075885 UP000183832 UP000000673 UP000030765 UP000092443 UP000092460 UP000027135 UP000053097 UP000076407 UP000075902 UP000092444 UP000075881 UP000092445 UP000076408 UP000075900 UP000005203 UP000002320 UP000242457 UP000053105 UP000078200 UP000075901 UP000075809 UP000078492 UP000078542 UP000002358 UP000036403 UP000078541 UP000005225 UP000005640
Interpro
ProteinModelPortal
A0A2W1BMT5
A0A2A4JQ51
A0A194QJS9
A0A1E1WQC2
S4NXX5
A0A212FB90
+ More
A0A0N1IGF8 A0A1E1W9Q7 B4N7Y5 A0A0K8VRT4 A0A0K8VPD3 B3P0V1 A0A0K8W618 B4PS46 Q9VF86 A0A0L0CRT5 A0A0A1WV85 B4QYJ0 B4HDX4 B3MSU7 A0A1I8N2U8 T1PAP2 A0A1I8PD57 A0A0K8TTK3 A0A034WBN8 A0A1W4VSU9 B4MBK1 A0A1L8EG10 W8BG72 Q299G1 A0A1L8E248 B4JS05 A0A3B0K0E1 A0A0M4F6R8 B4G546 B4KCY1 A0A1I8Q7R0 A0A182Q6L9 A0A182W596 A0A336MD05 A0A182JA65 A0A182H2K0 A0A182IAG5 A0A023ERE5 Q7QF04 A0A023ESK4 A0A2M4AAR5 A0A2M4BSL2 A0A182N0B6 A0A2M4BSL7 A0A2M3ZI14 A0A182MK31 A0A1S4FD93 A0A023EQA5 Q176F9 A0A336N1G9 A0A182VAA2 A0A2J7RFQ6 A0A182FJ31 A0A182PR26 A0A1J1HLP6 W5J988 A0A336MEI2 A0A084VLL9 A0A1Q3FAU4 A0A1A9Y8L6 A0A1B0BRF7 A0A067RI65 A0A1Q3FB23 A0A026W6X4 A0A182XPG6 A0A182TW90 D3TQA1 A0A182K9C2 A0A310SPG5 A0A1B0A2Q0 A0A1Y1L9G4 A0A182XY19 A0A182RQ73 A0A088AMU5 B0WE05 A0A2A3EF77 A0A0M8ZZH5 A0A1A9VLK6 A0A182T5D8 A0A151WMI2 A0A151IZ45 A0A195D497 E9IWS3 K7IMA4 A0A0J7NXF9 A0A195FXD6 A0A336LI78 H0XZM5 H7C3G9 A0A170ZF18
A0A0N1IGF8 A0A1E1W9Q7 B4N7Y5 A0A0K8VRT4 A0A0K8VPD3 B3P0V1 A0A0K8W618 B4PS46 Q9VF86 A0A0L0CRT5 A0A0A1WV85 B4QYJ0 B4HDX4 B3MSU7 A0A1I8N2U8 T1PAP2 A0A1I8PD57 A0A0K8TTK3 A0A034WBN8 A0A1W4VSU9 B4MBK1 A0A1L8EG10 W8BG72 Q299G1 A0A1L8E248 B4JS05 A0A3B0K0E1 A0A0M4F6R8 B4G546 B4KCY1 A0A1I8Q7R0 A0A182Q6L9 A0A182W596 A0A336MD05 A0A182JA65 A0A182H2K0 A0A182IAG5 A0A023ERE5 Q7QF04 A0A023ESK4 A0A2M4AAR5 A0A2M4BSL2 A0A182N0B6 A0A2M4BSL7 A0A2M3ZI14 A0A182MK31 A0A1S4FD93 A0A023EQA5 Q176F9 A0A336N1G9 A0A182VAA2 A0A2J7RFQ6 A0A182FJ31 A0A182PR26 A0A1J1HLP6 W5J988 A0A336MEI2 A0A084VLL9 A0A1Q3FAU4 A0A1A9Y8L6 A0A1B0BRF7 A0A067RI65 A0A1Q3FB23 A0A026W6X4 A0A182XPG6 A0A182TW90 D3TQA1 A0A182K9C2 A0A310SPG5 A0A1B0A2Q0 A0A1Y1L9G4 A0A182XY19 A0A182RQ73 A0A088AMU5 B0WE05 A0A2A3EF77 A0A0M8ZZH5 A0A1A9VLK6 A0A182T5D8 A0A151WMI2 A0A151IZ45 A0A195D497 E9IWS3 K7IMA4 A0A0J7NXF9 A0A195FXD6 A0A336LI78 H0XZM5 H7C3G9 A0A170ZF18
PDB
2CH6
E-value=1.44325e-58,
Score=573
Ontologies
PANTHER
Topology
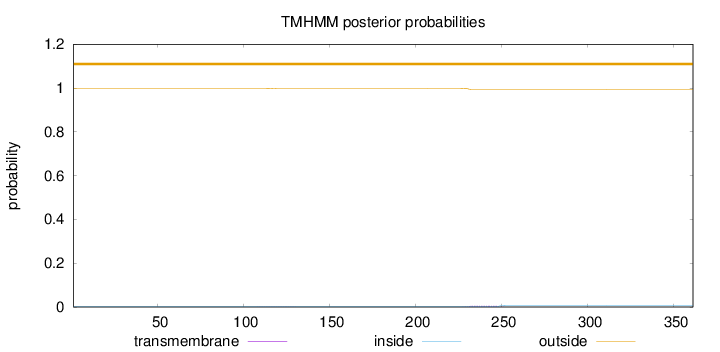
Length:
361
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10049
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00114
outside
1 - 361
Population Genetic Test Statistics
Pi
25.344468
Theta
19.831924
Tajima's D
0.162263
CLR
0.736046
CSRT
0.413679316034198
Interpretation
Uncertain
