Gene
KWMTBOMO15726
Annotation
PREDICTED:_sugar_transporter_4_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.959
Sequence
CDS
ATGGCAACCAGCGACCGGAAGGGAGTGCCCTCCGGTTCACTTCTCATCCACGTGGGGTCTACCATTACTGACAGACCCACACGTCTGACCACGGTGTGGCCACGCCGGCGTACCGTGGAAGAACAAAGTTCATCGACTTTGGAAGCCGAAGCACTGTCGAGGGATGTGGAACAGCGGGACAGTCAAAATGGCCACCCCAAAGTCCTCGTCCGCATGGAGACAATTCCGGATATCAAAAGGGATACATCCGGAATCACCACGCAATATGTGGGCACCGGTATAGTGAACCTTGGTGCTTTTGCCGCTGGAGTCTGTGTAGCGTGGTCATCATCAGCGTTGCCATTATTAATCACTGAATACGGAAAGGATTATACTACACCGATACCGGACTGGGCAGAGCAGAATTCGACTTATGATGAAAACTTTAAAACTCGTCTGGAGTTGTCAGATACCCAAGGATCCTGGGTGGCGTCACTCTTGTGTTTGGGAGCGGTGTTTGGCGCTGTACCCTCTGGCCTGATTTCCGAATACTTCGGTAGGAAAAAGACATTACTGTACTTGGCTTTACCACTGCTCGTCTCTTGGATTCTAGTCGCTTCGAGCCCAAACGTCTATGGTTTATATGTCGGAAGATTTGTGGGAGGAATAGCTGTGGGCGCATTCAGTGTTAGCATACCGCCTTACGTCGAAGACATAGCCGAAAAACATCTGCTCAAGACCTTAGCCAATTTCTATCACGTCGATTTTAATTGTGGCGTTTTGTTTGGGTACTTTATAGGTATAGTTGGAAACGTGTCTTGGCTTTCGGTTCTTTGCTCTTTAATACCCATCGCATTCTTCATTGCGTTTATATTCTTACCAGAATCTCCAACTTATCTGATGTCTCAGGGTAAATATAGTGAAGCCAAAGCAGCTCTGAGGTACTACCGAGGGATCGATAATGACATAGACGGAGAAATCAGAACATTGAGAGATTACTTGATGAACGCTGGCAAAAACAGGGTTTCATTCAAAGAGCTCTTCACTACGAGAGGCATGTTGAAACCTCTCTTGGTCTCTTTCTGTCTAATGATTTTTCAGCAAATGAGCGGGATTTATGCTGTTCTTTTCTATGCGAGAAAAATATTCAAGAATCTATCTGTCTCACTGAATCCGCCGAATGCTGCAATCATTCTAGGTTTCGGTCTGGTTTCTTCAACGTACTTCTCGACAATGCTGCTGAAGGTAGTCAGACGCAGAGTGTTGTTGATGACGTCATTTATAATGATGGCCTTAAACTTGGGCGGGTTGGCTATTTATTATCATTTGCAAGCTACGAATTTTTCATCCAATAATACGGGGGTGCCATTGTTCACTTTGTGTTTCTTCGTGATATTTTACGCCGCTGGAGCCGGTTCTATACCTTGGCTGATGCTTCGGGAGATATTTCCTCCACATGCAATCCGAAGGGCGACTGCCATAACAGCTGGTGTCCATTGGTTCCTGGCTTTCACTGTCACAAAATTATACCAGAATTTAGAAGATTTAGTGAAGCCCGGCTGGGCGTTTTGGCATTTTGCCGTTTCCTGCGTAGTTGGTACAGTGTTCGTTTACTTCTTTGTACCAGAAACTAAGGGTAGAAGCTTAGAAGACATACAAAACGAATTTGAAGGGATCCATAAGAAGAAAAGACACCGCCATGTGATTGAAGTCGAAAGTTTATCAGAGGTTTAA
Protein
MATSDRKGVPSGSLLIHVGSTITDRPTRLTTVWPRRRTVEEQSSSTLEAEALSRDVEQRDSQNGHPKVLVRMETIPDIKRDTSGITTQYVGTGIVNLGAFAAGVCVAWSSSALPLLITEYGKDYTTPIPDWAEQNSTYDENFKTRLELSDTQGSWVASLLCLGAVFGAVPSGLISEYFGRKKTLLYLALPLLVSWILVASSPNVYGLYVGRFVGGIAVGAFSVSIPPYVEDIAEKHLLKTLANFYHVDFNCGVLFGYFIGIVGNVSWLSVLCSLIPIAFFIAFIFLPESPTYLMSQGKYSEAKAALRYYRGIDNDIDGEIRTLRDYLMNAGKNRVSFKELFTTRGMLKPLLVSFCLMIFQQMSGIYAVLFYARKIFKNLSVSLNPPNAAIILGFGLVSSTYFSTMLLKVVRRRVLLMTSFIMMALNLGGLAIYYHLQATNFSSNNTGVPLFTLCFFVIFYAAGAGSIPWLMLREIFPPHAIRRATAITAGVHWFLAFTVTKLYQNLEDLVKPGWAFWHFAVSCVVGTVFVYFFVPETKGRSLEDIQNEFEGIHKKKRHRHVIEVESLSEV
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
D2XRA7
A0A2W1BHF1
A0A2H1V0P2
A0A2A4J550
A0A0N0PBS2
A0A0N0PA24
+ More
A0A2H1VDZ3 A0A212FED9 A0A2J7PYX6 A0A1W4WVK3 A0A2J7PIQ0 D6X0Y1 A0A1B6L0T0 A0A0K8UNM6 D4AHW9 A0A1B6MT94 A0A2P8YT21 A0A1B6E0I4 A0A1B6LM42 J3JVK7 A0A1B6M0Z3 A0A0V0G9V2 A0A088A2U8 A0A034VPA7 A0A067RCA0 A0A3L8DKM9 A0A0P4VHY9 D4AHX6 A0A023F661 N6TR17 U4U7Z5 T1HNI9 A0A0A1WTN4 A0A1I8M2U2 A0A224XPC2 T1PCW2 A0A067QP79 A0A1B6FS48 A0A069DV37 A0A0A1WG49 A0A2A3E6F6 A0A2J7QR87 A0A2J7QR88 A0A0L7QPI5 A0A2Z5U777 B3LWC6 E9IK55 A0A1Y0AWQ1 A0A1I8P360 A0A1I8P3B0 D3TR19 A0A0N8P1N8 A0A0Q9WK01 A0A2R7VR67 A0A2P8XWS8 A0A0Q9WVX8 A0A0J7N829 B4LBJ1 A0A0R3P5D7 A0A1Y1KAM2 A0A1Y1N7N8 A0A1Z1XG25 A0A1J1IWB4 A0A1S4FQN7 A0A232ERM9 A0A0K8VUC9 B5DQ85 A0A0L0CIX6 R4WIM6 B4IZM8 A0A0R3P588 B4LXB4 A0A034VDB8 A0A2H8TK86 D4AHW6 A0A1Y1NE39 A0A1I8NPR6 A0A1W4UVY7 A0A0Q9XE05 E0W2C6 A0A2S2R3Z9 A0A1I8NPR8 A0A224XN37 A0A0A1XNV7 A0A0Q9XD79 A0A336LQF3 A0A026WQ15 B4L0T8 A0A1I8MYG4 A0A0Q9WS19 A0A0M3QWT3 E9J4B0 A0A1Q3FK71 A0A1W4UHA6 T1PG78 J9JNG2 A0A194QPN7
A0A2H1VDZ3 A0A212FED9 A0A2J7PYX6 A0A1W4WVK3 A0A2J7PIQ0 D6X0Y1 A0A1B6L0T0 A0A0K8UNM6 D4AHW9 A0A1B6MT94 A0A2P8YT21 A0A1B6E0I4 A0A1B6LM42 J3JVK7 A0A1B6M0Z3 A0A0V0G9V2 A0A088A2U8 A0A034VPA7 A0A067RCA0 A0A3L8DKM9 A0A0P4VHY9 D4AHX6 A0A023F661 N6TR17 U4U7Z5 T1HNI9 A0A0A1WTN4 A0A1I8M2U2 A0A224XPC2 T1PCW2 A0A067QP79 A0A1B6FS48 A0A069DV37 A0A0A1WG49 A0A2A3E6F6 A0A2J7QR87 A0A2J7QR88 A0A0L7QPI5 A0A2Z5U777 B3LWC6 E9IK55 A0A1Y0AWQ1 A0A1I8P360 A0A1I8P3B0 D3TR19 A0A0N8P1N8 A0A0Q9WK01 A0A2R7VR67 A0A2P8XWS8 A0A0Q9WVX8 A0A0J7N829 B4LBJ1 A0A0R3P5D7 A0A1Y1KAM2 A0A1Y1N7N8 A0A1Z1XG25 A0A1J1IWB4 A0A1S4FQN7 A0A232ERM9 A0A0K8VUC9 B5DQ85 A0A0L0CIX6 R4WIM6 B4IZM8 A0A0R3P588 B4LXB4 A0A034VDB8 A0A2H8TK86 D4AHW6 A0A1Y1NE39 A0A1I8NPR6 A0A1W4UVY7 A0A0Q9XE05 E0W2C6 A0A2S2R3Z9 A0A1I8NPR8 A0A224XN37 A0A0A1XNV7 A0A0Q9XD79 A0A336LQF3 A0A026WQ15 B4L0T8 A0A1I8MYG4 A0A0Q9WS19 A0A0M3QWT3 E9J4B0 A0A1Q3FK71 A0A1W4UHA6 T1PG78 J9JNG2 A0A194QPN7
Pubmed
EMBL
GU244353
ADB08386.1
KZ150169
PZC72627.1
ODYU01000138
SOQ34407.1
+ More
NWSH01003195 PCG66808.1 KQ460820 KPJ12099.1 KQ459199 KPJ02956.1 ODYU01001852 SOQ38632.1 AGBW02008963 OWR52057.1 NEVH01020342 PNF21526.1 NEVH01025074 PNF16189.1 KQ971372 EFA10574.1 GEBQ01022823 JAT17154.1 GDHF01024196 GDHF01015258 GDHF01004851 JAI28118.1 JAI37056.1 JAI47463.1 AB549997 BAI83418.1 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 PYGN01000377 PSN47391.1 GEDC01005863 JAS31435.1 GEBQ01015167 JAT24810.1 BT127275 AEE62237.1 GEBQ01010386 JAT29591.1 GECL01001429 JAP04695.1 GAKP01015559 GAKP01015558 GAKP01015557 JAC43393.1 KK852731 KDR17493.1 QOIP01000007 RLU21005.1 GDKW01002266 JAI54329.1 AB550004 BAI83425.1 GBBI01002017 JAC16695.1 APGK01023894 KB740523 ENN80488.1 KB632063 ERL88448.1 ACPB03007656 GBXI01012080 JAD02212.1 GFTR01006577 JAW09849.1 KA646494 AFP61123.1 KK853179 KDR10227.1 GECZ01016734 JAS53035.1 GBGD01001322 JAC87567.1 GBXI01016924 JAC97367.1 KZ288353 PBC27327.1 NEVH01011922 PNF31099.1 PNF31098.1 KQ414813 KOC60547.1 AP017502 BBB06794.1 CH902617 EDV43759.1 GL763924 EFZ19033.1 KY921823 ART29412.1 EZ423871 ADD20147.1 KPU80444.1 CH940647 KRF85117.1 KK854007 PTY09150.1 PYGN01001230 PSN36458.1 KRF85118.1 LBMM01008561 KMQ88790.1 EDW70801.1 CH379069 KRT08322.1 GEZM01087689 JAV58533.1 GEZM01010696 JAV93901.1 KY350166 ARX98205.1 CVRI01000063 CRL04571.1 NNAY01002587 OXU20982.1 GDHF01009846 JAI42468.1 EDY73816.1 JRES01000423 KNC31409.1 AK417295 BAN20510.1 CH916366 EDV97803.1 KRT08321.1 CH940650 EDW67792.1 GAKP01019399 JAC39553.1 GFXV01002739 MBW14544.1 AB549994 BAI83415.1 GEZM01010698 GEZM01010697 JAV93897.1 CH933809 KRG06533.1 DS235877 EEB19782.1 GGMS01015536 MBY84739.1 GFTR01006576 JAW09850.1 GBXI01001611 JAD12681.1 KRG06534.1 UFQS01000107 UFQT01000107 SSW99705.1 SSX20085.1 KK107139 QOIP01000003 EZA57781.1 RLU25049.1 EDW19188.1 KRF83470.1 CP012525 ALC44659.1 GL768090 EFZ12343.1 GFDL01007076 JAV27969.1 KA647145 AFP61774.1 ABLF02034165 KQ461185 KPJ07467.1
NWSH01003195 PCG66808.1 KQ460820 KPJ12099.1 KQ459199 KPJ02956.1 ODYU01001852 SOQ38632.1 AGBW02008963 OWR52057.1 NEVH01020342 PNF21526.1 NEVH01025074 PNF16189.1 KQ971372 EFA10574.1 GEBQ01022823 JAT17154.1 GDHF01024196 GDHF01015258 GDHF01004851 JAI28118.1 JAI37056.1 JAI47463.1 AB549997 BAI83418.1 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 PYGN01000377 PSN47391.1 GEDC01005863 JAS31435.1 GEBQ01015167 JAT24810.1 BT127275 AEE62237.1 GEBQ01010386 JAT29591.1 GECL01001429 JAP04695.1 GAKP01015559 GAKP01015558 GAKP01015557 JAC43393.1 KK852731 KDR17493.1 QOIP01000007 RLU21005.1 GDKW01002266 JAI54329.1 AB550004 BAI83425.1 GBBI01002017 JAC16695.1 APGK01023894 KB740523 ENN80488.1 KB632063 ERL88448.1 ACPB03007656 GBXI01012080 JAD02212.1 GFTR01006577 JAW09849.1 KA646494 AFP61123.1 KK853179 KDR10227.1 GECZ01016734 JAS53035.1 GBGD01001322 JAC87567.1 GBXI01016924 JAC97367.1 KZ288353 PBC27327.1 NEVH01011922 PNF31099.1 PNF31098.1 KQ414813 KOC60547.1 AP017502 BBB06794.1 CH902617 EDV43759.1 GL763924 EFZ19033.1 KY921823 ART29412.1 EZ423871 ADD20147.1 KPU80444.1 CH940647 KRF85117.1 KK854007 PTY09150.1 PYGN01001230 PSN36458.1 KRF85118.1 LBMM01008561 KMQ88790.1 EDW70801.1 CH379069 KRT08322.1 GEZM01087689 JAV58533.1 GEZM01010696 JAV93901.1 KY350166 ARX98205.1 CVRI01000063 CRL04571.1 NNAY01002587 OXU20982.1 GDHF01009846 JAI42468.1 EDY73816.1 JRES01000423 KNC31409.1 AK417295 BAN20510.1 CH916366 EDV97803.1 KRT08321.1 CH940650 EDW67792.1 GAKP01019399 JAC39553.1 GFXV01002739 MBW14544.1 AB549994 BAI83415.1 GEZM01010698 GEZM01010697 JAV93897.1 CH933809 KRG06533.1 DS235877 EEB19782.1 GGMS01015536 MBY84739.1 GFTR01006576 JAW09850.1 GBXI01001611 JAD12681.1 KRG06534.1 UFQS01000107 UFQT01000107 SSW99705.1 SSX20085.1 KK107139 QOIP01000003 EZA57781.1 RLU25049.1 EDW19188.1 KRF83470.1 CP012525 ALC44659.1 GL768090 EFZ12343.1 GFDL01007076 JAV27969.1 KA647145 AFP61774.1 ABLF02034165 KQ461185 KPJ07467.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000235965
UP000192223
+ More
UP000007266 UP000245037 UP000005203 UP000027135 UP000279307 UP000019118 UP000030742 UP000015103 UP000095301 UP000242457 UP000053825 UP000007801 UP000095300 UP000008792 UP000036403 UP000001819 UP000183832 UP000215335 UP000037069 UP000001070 UP000192221 UP000009192 UP000009046 UP000053097 UP000092553 UP000007819
UP000007266 UP000245037 UP000005203 UP000027135 UP000279307 UP000019118 UP000030742 UP000015103 UP000095301 UP000242457 UP000053825 UP000007801 UP000095300 UP000008792 UP000036403 UP000001819 UP000183832 UP000215335 UP000037069 UP000001070 UP000192221 UP000009192 UP000009046 UP000053097 UP000092553 UP000007819
PRIDE
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
D2XRA7
A0A2W1BHF1
A0A2H1V0P2
A0A2A4J550
A0A0N0PBS2
A0A0N0PA24
+ More
A0A2H1VDZ3 A0A212FED9 A0A2J7PYX6 A0A1W4WVK3 A0A2J7PIQ0 D6X0Y1 A0A1B6L0T0 A0A0K8UNM6 D4AHW9 A0A1B6MT94 A0A2P8YT21 A0A1B6E0I4 A0A1B6LM42 J3JVK7 A0A1B6M0Z3 A0A0V0G9V2 A0A088A2U8 A0A034VPA7 A0A067RCA0 A0A3L8DKM9 A0A0P4VHY9 D4AHX6 A0A023F661 N6TR17 U4U7Z5 T1HNI9 A0A0A1WTN4 A0A1I8M2U2 A0A224XPC2 T1PCW2 A0A067QP79 A0A1B6FS48 A0A069DV37 A0A0A1WG49 A0A2A3E6F6 A0A2J7QR87 A0A2J7QR88 A0A0L7QPI5 A0A2Z5U777 B3LWC6 E9IK55 A0A1Y0AWQ1 A0A1I8P360 A0A1I8P3B0 D3TR19 A0A0N8P1N8 A0A0Q9WK01 A0A2R7VR67 A0A2P8XWS8 A0A0Q9WVX8 A0A0J7N829 B4LBJ1 A0A0R3P5D7 A0A1Y1KAM2 A0A1Y1N7N8 A0A1Z1XG25 A0A1J1IWB4 A0A1S4FQN7 A0A232ERM9 A0A0K8VUC9 B5DQ85 A0A0L0CIX6 R4WIM6 B4IZM8 A0A0R3P588 B4LXB4 A0A034VDB8 A0A2H8TK86 D4AHW6 A0A1Y1NE39 A0A1I8NPR6 A0A1W4UVY7 A0A0Q9XE05 E0W2C6 A0A2S2R3Z9 A0A1I8NPR8 A0A224XN37 A0A0A1XNV7 A0A0Q9XD79 A0A336LQF3 A0A026WQ15 B4L0T8 A0A1I8MYG4 A0A0Q9WS19 A0A0M3QWT3 E9J4B0 A0A1Q3FK71 A0A1W4UHA6 T1PG78 J9JNG2 A0A194QPN7
A0A2H1VDZ3 A0A212FED9 A0A2J7PYX6 A0A1W4WVK3 A0A2J7PIQ0 D6X0Y1 A0A1B6L0T0 A0A0K8UNM6 D4AHW9 A0A1B6MT94 A0A2P8YT21 A0A1B6E0I4 A0A1B6LM42 J3JVK7 A0A1B6M0Z3 A0A0V0G9V2 A0A088A2U8 A0A034VPA7 A0A067RCA0 A0A3L8DKM9 A0A0P4VHY9 D4AHX6 A0A023F661 N6TR17 U4U7Z5 T1HNI9 A0A0A1WTN4 A0A1I8M2U2 A0A224XPC2 T1PCW2 A0A067QP79 A0A1B6FS48 A0A069DV37 A0A0A1WG49 A0A2A3E6F6 A0A2J7QR87 A0A2J7QR88 A0A0L7QPI5 A0A2Z5U777 B3LWC6 E9IK55 A0A1Y0AWQ1 A0A1I8P360 A0A1I8P3B0 D3TR19 A0A0N8P1N8 A0A0Q9WK01 A0A2R7VR67 A0A2P8XWS8 A0A0Q9WVX8 A0A0J7N829 B4LBJ1 A0A0R3P5D7 A0A1Y1KAM2 A0A1Y1N7N8 A0A1Z1XG25 A0A1J1IWB4 A0A1S4FQN7 A0A232ERM9 A0A0K8VUC9 B5DQ85 A0A0L0CIX6 R4WIM6 B4IZM8 A0A0R3P588 B4LXB4 A0A034VDB8 A0A2H8TK86 D4AHW6 A0A1Y1NE39 A0A1I8NPR6 A0A1W4UVY7 A0A0Q9XE05 E0W2C6 A0A2S2R3Z9 A0A1I8NPR8 A0A224XN37 A0A0A1XNV7 A0A0Q9XD79 A0A336LQF3 A0A026WQ15 B4L0T8 A0A1I8MYG4 A0A0Q9WS19 A0A0M3QWT3 E9J4B0 A0A1Q3FK71 A0A1W4UHA6 T1PG78 J9JNG2 A0A194QPN7
PDB
4LDS
E-value=2.96164e-28,
Score=313
Ontologies
GO
Topology
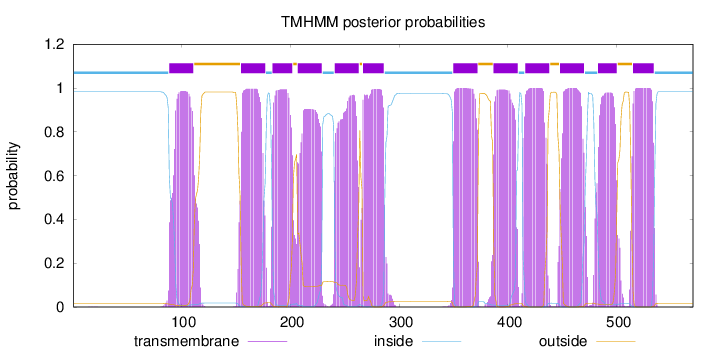
Length:
570
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
255.12733
Exp number, first 60 AAs:
0.00234
Total prob of N-in:
0.98330
inside
1 - 88
TMhelix
89 - 111
outside
112 - 154
TMhelix
155 - 177
inside
178 - 183
TMhelix
184 - 202
outside
203 - 206
TMhelix
207 - 229
inside
230 - 240
TMhelix
241 - 263
outside
264 - 266
TMhelix
267 - 286
inside
287 - 349
TMhelix
350 - 372
outside
373 - 386
TMhelix
387 - 409
inside
410 - 415
TMhelix
416 - 438
outside
439 - 447
TMhelix
448 - 470
inside
471 - 482
TMhelix
483 - 500
outside
501 - 514
TMhelix
515 - 534
inside
535 - 570
Population Genetic Test Statistics
Pi
206.127862
Theta
161.947009
Tajima's D
0.784911
CLR
0.003239
CSRT
0.602469876506175
Interpretation
Uncertain
