Gene
KWMTBOMO15720
Pre Gene Modal
BGIBMGA002312
Annotation
PREDICTED:_uncharacterized_protein_LOC106139274_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.471 Nuclear Reliability : 1.55
Sequence
CDS
ATGAACGAAAGTCGCGAACGCTTTCGACGCGAGAGGGAGCGAGAGAAGAACACGTACACGTCTCCCCGTCTCGCTCTACGCCGCGTGCTCCTGCTGGCCGAAGGGAGACAGTTCCGGGAAGCGGCTGCTATTGTATCAAGACTCGGACCGGGCGTGTTACAGAGCGTGGTCTCTGACTTGCCATTGGACTTGTTAATTGAAGCTTTACCTCATTCGGCGCACCTTATTGAGACGCTGCTAAATAGGCTCATATCGTTGGAGGTGTCTCCACGACCGGAGATACAATGCGAGTCGGTAGCCTGGAGGCTGGTCGGACTTCTCGGCGGAGATCAAGGGTCAGGACTTCGTGCCAGAACTTCGCGGCTCGCGTCTGCCCTGGTACACTATGCACCAGATTCAAGAGACGCTATTGATTCGAGGAGAAGACAACTCGACGCTGCTGTCCAAGGTCTTGGAACACACGGACTCACAGCAGACTCGTCAGGCACTCTGATATCCTTACATGTTGCCATGAAGAATGAGCTGCAAAGGCACGTAGAGGTCTACAAACAAGCATTACACAAGCTGGAGGAACTGTCACCGGTGACAGTAAACCAGGATCCAGCATCATCGTCCCACCAGCGTCTACTTGCTCTCTCCCACGCTGACGTGGAGCGTCGACTGATTGACAACAAGTCCCTGCTGACCATCGTTGATAAGCCAGCCTTAAGGCAGCTACCCACATTGGTCGCATCTTTAGCAGCACGGGTGGAGAGCGACAAAGCAGTGCTAGCCTGCATTGGACAGATCAAGAGAACTGATCCGACTTTAGATTTGAGCGATACGAGACCGGTAGCGGGTGTGCTTATGAGCTACTCCCGGGGTTGTGCAGCCGTCCTCAGTCAAATGTCCGAAGCAGAAGCTACAGGAGGACAGTCGGTCGTGCCTCGGTCACCGACGGACTCTGCCAGCGATGGATATCATTCCGACAGTGACGACTCACATCAGCATCCGGATAGGAGCAAATTGGTTTCTGAGTATGCGTCAGTGTGGGCGCGGGCCGCAGCAGAGGCGCTCCCGGCATTGGCGGCGCTGGAGCCACTGCGACCGACGCCGCACTTGAAGTACAAGATCGTCTTCTCTGTTGTTGTTCTCTCTTTCCGAGCAGTAATAAGTCTTCGTGATCGTCGCTTGTCCGAAGTGAAGGCTCTACTCGGAGTGAACGAAGAGACTAAAGCCAACGAAGCCACGGTCTCGCGGCTTATTGTTGCAGCGACACGGATGCTACAGGAAACAGCTCACAGCTTCCCTCTCGGTGATGCCGAGAGAACGGTTGTGAATCAGGTGCTGAATACTCTCCGGGAGTACCCTTGCCTGGCCTCATGCGCGGCTCTACACGCGCTGGTCGCGGCCGCCACGCGCGCAGCTTGGACCATCAGCTTGCACTCACCACCACTTAGAATCGACACCGACTTCACTCCTGTGTTAATGAACCCAGAGAAGCATGTAAGGTTTTCGGCGGGAGAGCTAAAGGACAGACGCTCTGACCTCATCAAATCTTTCGTGTGGCCGGCGTTAATGGATGGCAATCGATGCGTGTTCAGAGCCGTTGTACTTACATAG
Protein
MNESRERFRREREREKNTYTSPRLALRRVLLLAEGRQFREAAAIVSRLGPGVLQSVVSDLPLDLLIEALPHSAHLIETLLNRLISLEVSPRPEIQCESVAWRLVGLLGGDQGSGLRARTSRLASALVHYAPDSRDAIDSRRRQLDAAVQGLGTHGLTADSSGTLISLHVAMKNELQRHVEVYKQALHKLEELSPVTVNQDPASSSHQRLLALSHADVERRLIDNKSLLTIVDKPALRQLPTLVASLAARVESDKAVLACIGQIKRTDPTLDLSDTRPVAGVLMSYSRGCAAVLSQMSEAEATGGQSVVPRSPTDSASDGYHSDSDDSHQHPDRSKLVSEYASVWARAAAEALPALAALEPLRPTPHLKYKIVFSVVVLSFRAVISLRDRRLSEVKALLGVNEETKANEATVSRLIVAATRMLQETAHSFPLGDAERTVVNQVLNTLREYPCLASCAALHALVAAATRAAWTISLHSPPLRIDTDFTPVLMNPEKHVRFSAGELKDRRSDLIKSFVWPALMDGNRCVFRAVVLT
Summary
Uniprot
A0A2W1BDC3
A0A212FEA8
A0A0N1PEF1
A0A2A4JTF7
A0A2A4JT82
A0A0N1PIF0
+ More
A0A182INR7 A0A182H0S6 A0A182M7M0 A0A182VZC9 A0A182FQ38 A0A182NS34 A0A182TIS6 A0A182UWX7 A0A182WSI7 A0A182L9R8 A0A182HLD9 A0A182PGM9 A0A182YAJ1 Q7PXW1 A0A182R612 A0A182QLQ9 B4M6J7 B4K818 A0A1B0CUS3 B3LZC5 W5J702 B4JET4 A0A034W3A6 A0A0A1X2Q3 A0A0K8WAQ5 A0A0L0BZK5 A0A1A9VPW3 B4NGS5 A0A1I8MMQ8 A0A1I8PUT5 W8BL64 W8AZ17 A0A1I8PUQ4 A0A1I8PUQ6 B4HZM8 Q29C20 A0A3B0K774 Q9VAA0 A0A1B0DAC8 A0A3B0K9A6 B4PLT2 A0A1W4VRJ2 A0A1Z1CH18 B4R1J0 B3P7X8 A0A0M3QXN6 A0A1A9ZF88 A0A182K9E6 A0A1S4F2C8 A0A1B0GGG9 B4GNQ4 A0A084VYV6 A0A1A9XNU9 A0A1B0B1J1 D6X0V5 A0A1J1HX41 A0A151WQQ7 A0A026WAB5 A0A195E3Z7 E9ICA8 A0A088AEZ8 E2A2Z3 K7INX9 A0A232FHK1 A0A195FGZ7 A0A195BQS5 F4X4B0 A0A067RKK6 A0A158NXL0 E2BH28 A0A2A3EEJ3 A0A154PFI7 A0A0L7QKP7
A0A182INR7 A0A182H0S6 A0A182M7M0 A0A182VZC9 A0A182FQ38 A0A182NS34 A0A182TIS6 A0A182UWX7 A0A182WSI7 A0A182L9R8 A0A182HLD9 A0A182PGM9 A0A182YAJ1 Q7PXW1 A0A182R612 A0A182QLQ9 B4M6J7 B4K818 A0A1B0CUS3 B3LZC5 W5J702 B4JET4 A0A034W3A6 A0A0A1X2Q3 A0A0K8WAQ5 A0A0L0BZK5 A0A1A9VPW3 B4NGS5 A0A1I8MMQ8 A0A1I8PUT5 W8BL64 W8AZ17 A0A1I8PUQ4 A0A1I8PUQ6 B4HZM8 Q29C20 A0A3B0K774 Q9VAA0 A0A1B0DAC8 A0A3B0K9A6 B4PLT2 A0A1W4VRJ2 A0A1Z1CH18 B4R1J0 B3P7X8 A0A0M3QXN6 A0A1A9ZF88 A0A182K9E6 A0A1S4F2C8 A0A1B0GGG9 B4GNQ4 A0A084VYV6 A0A1A9XNU9 A0A1B0B1J1 D6X0V5 A0A1J1HX41 A0A151WQQ7 A0A026WAB5 A0A195E3Z7 E9ICA8 A0A088AEZ8 E2A2Z3 K7INX9 A0A232FHK1 A0A195FGZ7 A0A195BQS5 F4X4B0 A0A067RKK6 A0A158NXL0 E2BH28 A0A2A3EEJ3 A0A154PFI7 A0A0L7QKP7
Pubmed
28756777
22118469
26354079
26483478
20966253
25244985
+ More
12364791 14747013 17210077 17994087 18057021 20920257 23761445 25348373 25830018 26108605 25315136 24495485 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588 18362917 19820115 24508170 30249741 21282665 20798317 20075255 28648823 21719571 24845553 21347285
12364791 14747013 17210077 17994087 18057021 20920257 23761445 25348373 25830018 26108605 25315136 24495485 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588 18362917 19820115 24508170 30249741 21282665 20798317 20075255 28648823 21719571 24845553 21347285
EMBL
KZ150119
PZC73229.1
AGBW02008963
OWR52060.1
KQ459199
KPJ02960.1
+ More
NWSH01000705 PCG74672.1 PCG74673.1 KQ460820 KPJ12103.1 AXCP01007616 JXUM01021287 JXUM01021288 JXUM01021289 JXUM01021290 JXUM01022761 JXUM01022762 KQ560640 KQ560596 KXJ81477.1 KXJ81704.1 AXCM01001257 APCN01000894 AAAB01008987 EAA00981.5 AXCN02000266 CH940652 EDW59273.1 CH933806 EDW14352.1 KRG00973.1 AJWK01029613 CH902617 EDV44104.2 ADMH02002100 ETN59178.1 CH916369 EDV93215.1 GAKP01008866 JAC50086.1 GBXI01009631 GBXI01008925 JAD04661.1 JAD05367.1 GDHF01028388 GDHF01016708 GDHF01004137 JAI23926.1 JAI35606.1 JAI48177.1 JRES01001112 KNC25465.1 CH964272 EDW84422.1 GAMC01012509 JAB94046.1 GAMC01012510 JAB94045.1 CH480819 EDW53485.1 CM000070 EAL26826.3 OUUW01000041 SPP89944.1 AE014297 BT099876 AAF57014.3 ACX30038.1 AJVK01013270 SPP89943.1 CM000160 EDW99069.2 KRK04643.1 API64951.1 CM000364 EDX14978.1 CH954182 EDV53105.1 CP012526 ALC46243.1 CCAG010012084 CH479186 EDW39380.1 ATLV01018496 ATLV01018497 ATLV01018498 KE525235 KFB43150.1 JXJN01007187 KQ971372 EFA09543.2 CVRI01000023 CRK92090.1 KQ982815 KYQ50242.1 KK107323 QOIP01000002 EZA52586.1 RLU25876.1 KQ979685 KYN19806.1 GL762231 EFZ21777.1 GL436239 EFN72201.1 NNAY01000235 OXU29817.1 KQ981606 KYN39512.1 KQ976423 KYM88921.1 GL888633 EGI58706.1 KK852458 KDR23548.1 ADTU01000471 ADTU01000472 ADTU01000473 ADTU01000474 GL448268 EFN84949.1 KZ288287 PBC29431.1 KQ434892 KZC10572.1 KQ414940 KOC59218.1
NWSH01000705 PCG74672.1 PCG74673.1 KQ460820 KPJ12103.1 AXCP01007616 JXUM01021287 JXUM01021288 JXUM01021289 JXUM01021290 JXUM01022761 JXUM01022762 KQ560640 KQ560596 KXJ81477.1 KXJ81704.1 AXCM01001257 APCN01000894 AAAB01008987 EAA00981.5 AXCN02000266 CH940652 EDW59273.1 CH933806 EDW14352.1 KRG00973.1 AJWK01029613 CH902617 EDV44104.2 ADMH02002100 ETN59178.1 CH916369 EDV93215.1 GAKP01008866 JAC50086.1 GBXI01009631 GBXI01008925 JAD04661.1 JAD05367.1 GDHF01028388 GDHF01016708 GDHF01004137 JAI23926.1 JAI35606.1 JAI48177.1 JRES01001112 KNC25465.1 CH964272 EDW84422.1 GAMC01012509 JAB94046.1 GAMC01012510 JAB94045.1 CH480819 EDW53485.1 CM000070 EAL26826.3 OUUW01000041 SPP89944.1 AE014297 BT099876 AAF57014.3 ACX30038.1 AJVK01013270 SPP89943.1 CM000160 EDW99069.2 KRK04643.1 API64951.1 CM000364 EDX14978.1 CH954182 EDV53105.1 CP012526 ALC46243.1 CCAG010012084 CH479186 EDW39380.1 ATLV01018496 ATLV01018497 ATLV01018498 KE525235 KFB43150.1 JXJN01007187 KQ971372 EFA09543.2 CVRI01000023 CRK92090.1 KQ982815 KYQ50242.1 KK107323 QOIP01000002 EZA52586.1 RLU25876.1 KQ979685 KYN19806.1 GL762231 EFZ21777.1 GL436239 EFN72201.1 NNAY01000235 OXU29817.1 KQ981606 KYN39512.1 KQ976423 KYM88921.1 GL888633 EGI58706.1 KK852458 KDR23548.1 ADTU01000471 ADTU01000472 ADTU01000473 ADTU01000474 GL448268 EFN84949.1 KZ288287 PBC29431.1 KQ434892 KZC10572.1 KQ414940 KOC59218.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000053240
UP000075880
UP000069940
+ More
UP000249989 UP000075883 UP000075920 UP000069272 UP000075884 UP000075902 UP000075903 UP000076407 UP000075882 UP000075840 UP000075885 UP000076408 UP000007062 UP000075900 UP000075886 UP000008792 UP000009192 UP000092461 UP000007801 UP000000673 UP000001070 UP000037069 UP000078200 UP000007798 UP000095301 UP000095300 UP000001292 UP000001819 UP000268350 UP000000803 UP000092462 UP000002282 UP000192221 UP000000304 UP000008711 UP000092553 UP000092445 UP000075881 UP000092444 UP000008744 UP000030765 UP000092443 UP000092460 UP000007266 UP000183832 UP000075809 UP000053097 UP000279307 UP000078492 UP000005203 UP000000311 UP000002358 UP000215335 UP000078541 UP000078540 UP000007755 UP000027135 UP000005205 UP000008237 UP000242457 UP000076502 UP000053825
UP000249989 UP000075883 UP000075920 UP000069272 UP000075884 UP000075902 UP000075903 UP000076407 UP000075882 UP000075840 UP000075885 UP000076408 UP000007062 UP000075900 UP000075886 UP000008792 UP000009192 UP000092461 UP000007801 UP000000673 UP000001070 UP000037069 UP000078200 UP000007798 UP000095301 UP000095300 UP000001292 UP000001819 UP000268350 UP000000803 UP000092462 UP000002282 UP000192221 UP000000304 UP000008711 UP000092553 UP000092445 UP000075881 UP000092444 UP000008744 UP000030765 UP000092443 UP000092460 UP000007266 UP000183832 UP000075809 UP000053097 UP000279307 UP000078492 UP000005203 UP000000311 UP000002358 UP000215335 UP000078541 UP000078540 UP000007755 UP000027135 UP000005205 UP000008237 UP000242457 UP000076502 UP000053825
PRIDE
Pfam
PF16026 MIEAP
ProteinModelPortal
A0A2W1BDC3
A0A212FEA8
A0A0N1PEF1
A0A2A4JTF7
A0A2A4JT82
A0A0N1PIF0
+ More
A0A182INR7 A0A182H0S6 A0A182M7M0 A0A182VZC9 A0A182FQ38 A0A182NS34 A0A182TIS6 A0A182UWX7 A0A182WSI7 A0A182L9R8 A0A182HLD9 A0A182PGM9 A0A182YAJ1 Q7PXW1 A0A182R612 A0A182QLQ9 B4M6J7 B4K818 A0A1B0CUS3 B3LZC5 W5J702 B4JET4 A0A034W3A6 A0A0A1X2Q3 A0A0K8WAQ5 A0A0L0BZK5 A0A1A9VPW3 B4NGS5 A0A1I8MMQ8 A0A1I8PUT5 W8BL64 W8AZ17 A0A1I8PUQ4 A0A1I8PUQ6 B4HZM8 Q29C20 A0A3B0K774 Q9VAA0 A0A1B0DAC8 A0A3B0K9A6 B4PLT2 A0A1W4VRJ2 A0A1Z1CH18 B4R1J0 B3P7X8 A0A0M3QXN6 A0A1A9ZF88 A0A182K9E6 A0A1S4F2C8 A0A1B0GGG9 B4GNQ4 A0A084VYV6 A0A1A9XNU9 A0A1B0B1J1 D6X0V5 A0A1J1HX41 A0A151WQQ7 A0A026WAB5 A0A195E3Z7 E9ICA8 A0A088AEZ8 E2A2Z3 K7INX9 A0A232FHK1 A0A195FGZ7 A0A195BQS5 F4X4B0 A0A067RKK6 A0A158NXL0 E2BH28 A0A2A3EEJ3 A0A154PFI7 A0A0L7QKP7
A0A182INR7 A0A182H0S6 A0A182M7M0 A0A182VZC9 A0A182FQ38 A0A182NS34 A0A182TIS6 A0A182UWX7 A0A182WSI7 A0A182L9R8 A0A182HLD9 A0A182PGM9 A0A182YAJ1 Q7PXW1 A0A182R612 A0A182QLQ9 B4M6J7 B4K818 A0A1B0CUS3 B3LZC5 W5J702 B4JET4 A0A034W3A6 A0A0A1X2Q3 A0A0K8WAQ5 A0A0L0BZK5 A0A1A9VPW3 B4NGS5 A0A1I8MMQ8 A0A1I8PUT5 W8BL64 W8AZ17 A0A1I8PUQ4 A0A1I8PUQ6 B4HZM8 Q29C20 A0A3B0K774 Q9VAA0 A0A1B0DAC8 A0A3B0K9A6 B4PLT2 A0A1W4VRJ2 A0A1Z1CH18 B4R1J0 B3P7X8 A0A0M3QXN6 A0A1A9ZF88 A0A182K9E6 A0A1S4F2C8 A0A1B0GGG9 B4GNQ4 A0A084VYV6 A0A1A9XNU9 A0A1B0B1J1 D6X0V5 A0A1J1HX41 A0A151WQQ7 A0A026WAB5 A0A195E3Z7 E9ICA8 A0A088AEZ8 E2A2Z3 K7INX9 A0A232FHK1 A0A195FGZ7 A0A195BQS5 F4X4B0 A0A067RKK6 A0A158NXL0 E2BH28 A0A2A3EEJ3 A0A154PFI7 A0A0L7QKP7
Ontologies
PANTHER
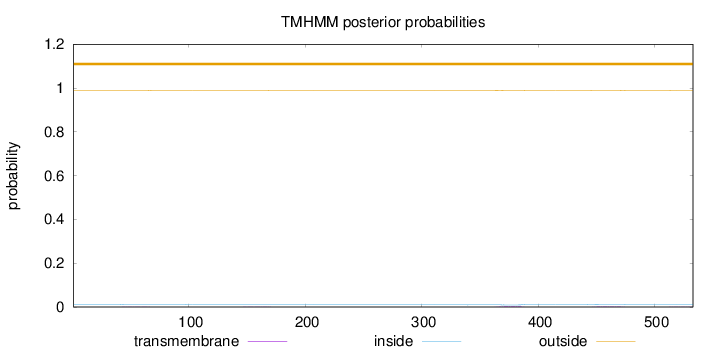
Topology
Length:
533
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1377
Exp number, first 60 AAs:
0.00966
Total prob of N-in:
0.01118
outside
1 - 533
Population Genetic Test Statistics
Pi
222.075306
Theta
157.947442
Tajima's D
1.826313
CLR
0.234165
CSRT
0.853957302134893
Interpretation
Uncertain
