Gene
KWMTBOMO15716
Pre Gene Modal
BGIBMGA002260
Annotation
PREDICTED:_protein_C12orf4_homolog_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.621
Sequence
CDS
ATGATTTCAGAAACAATTCAAAATACAACAAAAACCTTTAAATATTCATTTCCAACATGCACGAACGAAAATGTCTTGTTCAATCTAGAAGTTCCTGTGGATATTCCGTATGATGGTTCCACGAGAGAGCTCGTACAAAGAGTTATAAATATGTTTCACATTCCAGTATATTTAGAAGAAGAATTAAACGATAGACTATCAGAATTTGTAGCTGACGAAACAAAAAGATTTCACAACGAAAGAGATGAAGAATTATTGAACAAACTAAAGAATAATGAAATAAACTTAGATGATGTTGTTCAAAAATGGGAAAAGCTCTTTAAAGAGAACGTTGTGGAATTCGCAGAACAAAAAGGAACCCCAGATGAAGAGGTATTTGCAGTAGCATACCACAAATTAGTTCACTCACCAGCTCTGGAGACCATTCTACAAGTAGAAAACTCCTATGCCAAAACCGTTATGAATACCTTAAAGAATAGAGATGATGATATCGTAAAACTGACACAAAAACAAACTGAAGAAATGGAAGAAAAAGTACGTCTTCTGAACACTTCAACCACAGAAGTTGAGATCAATGAATTAGCCACCCAGCAGTTTGAAGAGCAAGCCTTAGTTTCAGGCCGATGGGGATCTCAGCTGGACGCCTTGTGTCAAACGCAAAGAACACATCACAGAGCATGGCTAATGGCGACTTTGCAGGAGTACCAGACCACATCAGCCTTGCATACACCAAGCAATTCTCCGCTGGGTATATTCTCGCCGTCTCCATGCTCATCCCTACCCGAGGCTCCAACCCGACCTCGTCTTGAAGAAAGCTTCACCATACACCTCGGAAGTCAACTCAAACAGACGCACAACATTCGCATTGTCGCTATGGACCTGCTCGACCTGTGCAATGTTGATCATGACAACCCTACAGAATCGTCACGTCGCCTCCAAACTTCCCTATCGTTGTATTCATCGGAACTATCAGCGGTGATTCTCCTCAGCGAGTCTCCCCCTGAACGACCCTCGCCCCCCACCACACAGCTAGTGGACGCGGCCAAGTCCTCGACGGAGTATCACTTCCACGACGTCGAGTATCAATTGCGGGAAATCAGCAAGAAAGTCAAGGAGCCAGTCGAACGCCGTAACACTCAGCGCAGCAAAGAACAAAACGACAGCAATAACACGCAAACCCACAACAACCCATTTAGGAACAGATGGAGACAAAACCTGCAGACTGGAGACATATACCTGACTAAGCACAGCAACTTGGCTGACGTGCACGTTGTCTTCCATCTTGTTGCCGACGACTCTTCTCTGAGATCCGGTGATATCACGTCTCGTCATCCGGTAATCCTCGGCTTACGTAACATCCTGAAAGCGGCTTGCAGTAATGATATCACCAGTATATCGCTGCCTCTGCTGTTGCGACACGAATTATCAGAGGAAATGACGGCAGCGTGGTGCATCCGTCGTGCGGAATTGATCCTGAAATGCGTCAAAGGGTTCATGTTGGAGGCGGCCGGGTGGGGCGGGGCTGAACTCCGCACCCTACAGTTCGCAGTACCAACGAACACCGCGCCAGAAGTGTTTTCGACTCTCGCTAATCTACTGCCCTCTATATTCAGAATATCTAATCCGGTCAAGTTTAAAGCGAACTCTTAA
Protein
MISETIQNTTKTFKYSFPTCTNENVLFNLEVPVDIPYDGSTRELVQRVINMFHIPVYLEEELNDRLSEFVADETKRFHNERDEELLNKLKNNEINLDDVVQKWEKLFKENVVEFAEQKGTPDEEVFAVAYHKLVHSPALETILQVENSYAKTVMNTLKNRDDDIVKLTQKQTEEMEEKVRLLNTSTTEVEINELATQQFEEQALVSGRWGSQLDALCQTQRTHHRAWLMATLQEYQTTSALHTPSNSPLGIFSPSPCSSLPEAPTRPRLEESFTIHLGSQLKQTHNIRIVAMDLLDLCNVDHDNPTESSRRLQTSLSLYSSELSAVILLSESPPERPSPPTTQLVDAAKSSTEYHFHDVEYQLREISKKVKEPVERRNTQRSKEQNDSNNTQTHNNPFRNRWRQNLQTGDIYLTKHSNLADVHVVFHLVADDSSLRSGDITSRHPVILGLRNILKAACSNDITSISLPLLLRHELSEEMTAAWCIRRAELILKCVKGFMLEAAGWGGAELRTLQFAVPTNTAPEVFSTLANLLPSIFRISNPVKFKANS
Summary
Uniprot
H9IYC7
A0A2W1BHN7
A0A2A4J411
A0A0N1PG23
A0A0N1PFL8
A0A212FE71
+ More
A0A067R148 A0A2J7R583 F4X8L9 A0A151WF79 A0A232F201 A0A3L8DVW3 A0A026W6T0 E2A1I7 E2BIG5 A0A154P745 A0A158NBK3 A0A195DRE1 A0A0C9PVL5 A0A0J7LA42 A0A0L7QRM3 A0A2A3ETN7 A0A088AE66 A0A195B9R9 A0A1B6G7P0 E9J9G1 A0A1B6L9E9 A0A195C6G4 A0A158NC14 A0A0C9RB08 D6W6Q1 J9JV81 A0A2H8TL98 A0A2S2P6I4 A0A2S2Q0V7 N6U5G4 A0A2J7R590 A0A0A9Z135 A0A1Y1N9I2 E0VWS5 K1QEZ2 A0A023F0I4 A0A0V0GCB7 A0A224XMZ4 A0A069DUJ6 V3ZTG6 T1JFI6 A0A2C9JQK1 A0A087U8J7 A0A1I8NN21 A0A1I8M679 A0A0P4VNU8 B4JUA0 R7VA97 A0A0L0BYE2 A0A0L8G0S4 A0A3Q3BR28 A0A3Q1FR27 A0A3P8RKM3 A0A3B0K724 Q29BI8 A0A3B4GSI6 I3JVM2 A0A3S1BPI5 B4K8E1 A0A3P8YAG8 A0A2I4DBK6 A0A3Q3VPF0 A0A2I4DBL8 A0A1E1XM36 B4LWP0 A0A3P8PI24 A0A3P9CHS7 A0A3Q0R6R2 A0A293M2Y3 A0JMN9 H2TJU3 A0A0F8AES9 A0A0K8U0J3 A0A146TQ48 A0A3B5Q704 A0A3B4ZYT3 A0A3P9MWE8 B4GP02 A0A3Q1IR09 A0A3Q2FRI4 A0A3Q3E058 A0A1S3K0H4 A0A1A7WQA9 B3P5S0 A0A210QSS9 A0A1S3MK28 A0A3B4VIU8 A0A087XLF2 A0A2I4DBJ8 A0A1W4W2J0 A0A2R5LF48 A0A3B3DY19
A0A067R148 A0A2J7R583 F4X8L9 A0A151WF79 A0A232F201 A0A3L8DVW3 A0A026W6T0 E2A1I7 E2BIG5 A0A154P745 A0A158NBK3 A0A195DRE1 A0A0C9PVL5 A0A0J7LA42 A0A0L7QRM3 A0A2A3ETN7 A0A088AE66 A0A195B9R9 A0A1B6G7P0 E9J9G1 A0A1B6L9E9 A0A195C6G4 A0A158NC14 A0A0C9RB08 D6W6Q1 J9JV81 A0A2H8TL98 A0A2S2P6I4 A0A2S2Q0V7 N6U5G4 A0A2J7R590 A0A0A9Z135 A0A1Y1N9I2 E0VWS5 K1QEZ2 A0A023F0I4 A0A0V0GCB7 A0A224XMZ4 A0A069DUJ6 V3ZTG6 T1JFI6 A0A2C9JQK1 A0A087U8J7 A0A1I8NN21 A0A1I8M679 A0A0P4VNU8 B4JUA0 R7VA97 A0A0L0BYE2 A0A0L8G0S4 A0A3Q3BR28 A0A3Q1FR27 A0A3P8RKM3 A0A3B0K724 Q29BI8 A0A3B4GSI6 I3JVM2 A0A3S1BPI5 B4K8E1 A0A3P8YAG8 A0A2I4DBK6 A0A3Q3VPF0 A0A2I4DBL8 A0A1E1XM36 B4LWP0 A0A3P8PI24 A0A3P9CHS7 A0A3Q0R6R2 A0A293M2Y3 A0JMN9 H2TJU3 A0A0F8AES9 A0A0K8U0J3 A0A146TQ48 A0A3B5Q704 A0A3B4ZYT3 A0A3P9MWE8 B4GP02 A0A3Q1IR09 A0A3Q2FRI4 A0A3Q3E058 A0A1S3K0H4 A0A1A7WQA9 B3P5S0 A0A210QSS9 A0A1S3MK28 A0A3B4VIU8 A0A087XLF2 A0A2I4DBJ8 A0A1W4W2J0 A0A2R5LF48 A0A3B3DY19
Pubmed
19121390
28756777
26354079
22118469
24845553
21719571
+ More
28648823 30249741 24508170 20798317 21347285 21282665 18362917 19820115 23537049 25401762 26823975 28004739 20566863 22992520 25474469 26334808 23254933 15562597 25315136 27129103 17994087 26108605 15632085 25186727 18057021 25069045 29209593 23594743 21551351 25835551 23542700 28812685 29451363
28648823 30249741 24508170 20798317 21347285 21282665 18362917 19820115 23537049 25401762 26823975 28004739 20566863 22992520 25474469 26334808 23254933 15562597 25315136 27129103 17994087 26108605 15632085 25186727 18057021 25069045 29209593 23594743 21551351 25835551 23542700 28812685 29451363
EMBL
BABH01020639
KZ150119
PZC73225.1
NWSH01003383
PCG66418.1
KQ459199
+ More
KPJ02967.1 KQ460820 KPJ12108.1 AGBW02008963 OWR52065.1 KK852818 KDR15654.1 NEVH01007392 PNF35998.1 GL888932 EGI57288.1 KQ983227 KYQ46492.1 NNAY01001183 OXU24826.1 QOIP01000004 RLU24235.1 KK107372 EZA51765.1 GL435766 EFN72660.1 GL448504 EFN84553.1 KQ434827 KZC07653.1 ADTU01011144 KQ980581 KYN15377.1 GBYB01005498 JAG75265.1 LBMM01000076 KMR05056.1 KQ414775 KOC61290.1 KZ288192 PBC34411.1 KQ976540 KYM81276.1 GECZ01025642 GECZ01011335 JAS44127.1 JAS58434.1 GL769313 EFZ10531.1 GEBQ01019654 JAT20323.1 KQ978219 KYM96432.1 ADTU01011145 ADTU01011146 GBYB01005500 JAG75267.1 KQ971307 EFA10962.1 ABLF02033606 GFXV01003121 MBW14926.1 GGMR01012444 MBY25063.1 GGMS01002101 MBY71304.1 APGK01039126 APGK01039127 KB740967 KB632399 ENN76845.1 ERL94666.1 PNF35997.1 GBHO01031685 GBHO01005456 GBHO01005450 GDHC01007069 JAG11919.1 JAG38148.1 JAG38154.1 JAQ11560.1 GEZM01011376 JAV93540.1 DS235824 EEB17831.1 JH817657 EKC29664.1 GBBI01003730 JAC14982.1 GECL01000494 JAP05630.1 GFTR01007035 JAW09391.1 GBGD01001096 JAC87793.1 KB203566 ESO84206.1 JH432169 KK118720 KFM73686.1 GDKW01001948 JAI54647.1 CH916374 EDV91070.1 AMQN01004487 KB293638 ELU15738.1 JRES01001160 KNC25040.1 KQ424993 KOF70180.1 OUUW01000005 SPP80801.1 CM000070 EAL27010.2 AERX01017131 RQTK01000083 RUS88413.1 CH933806 EDW16523.1 KRG02179.1 GFAA01003120 JAU00315.1 CH940650 EDW67705.1 GFWV01010543 MAA35272.1 CR396584 BC125950 AAI25951.1 KQ042670 KKF12078.1 GDHF01032253 GDHF01029832 GDHF01023829 GDHF01021543 GDHF01012690 GDHF01010437 JAI20061.1 JAI22482.1 JAI28485.1 JAI30771.1 JAI39624.1 JAI41877.1 GCES01091339 JAQ94983.1 CH479186 EDW38885.1 HADW01006505 HADX01009383 SBP07905.1 CH954182 EDV53320.1 NEDP02002059 OWF51817.1 AYCK01018557 AYCK01018558 GGLE01004024 MBY08150.1
KPJ02967.1 KQ460820 KPJ12108.1 AGBW02008963 OWR52065.1 KK852818 KDR15654.1 NEVH01007392 PNF35998.1 GL888932 EGI57288.1 KQ983227 KYQ46492.1 NNAY01001183 OXU24826.1 QOIP01000004 RLU24235.1 KK107372 EZA51765.1 GL435766 EFN72660.1 GL448504 EFN84553.1 KQ434827 KZC07653.1 ADTU01011144 KQ980581 KYN15377.1 GBYB01005498 JAG75265.1 LBMM01000076 KMR05056.1 KQ414775 KOC61290.1 KZ288192 PBC34411.1 KQ976540 KYM81276.1 GECZ01025642 GECZ01011335 JAS44127.1 JAS58434.1 GL769313 EFZ10531.1 GEBQ01019654 JAT20323.1 KQ978219 KYM96432.1 ADTU01011145 ADTU01011146 GBYB01005500 JAG75267.1 KQ971307 EFA10962.1 ABLF02033606 GFXV01003121 MBW14926.1 GGMR01012444 MBY25063.1 GGMS01002101 MBY71304.1 APGK01039126 APGK01039127 KB740967 KB632399 ENN76845.1 ERL94666.1 PNF35997.1 GBHO01031685 GBHO01005456 GBHO01005450 GDHC01007069 JAG11919.1 JAG38148.1 JAG38154.1 JAQ11560.1 GEZM01011376 JAV93540.1 DS235824 EEB17831.1 JH817657 EKC29664.1 GBBI01003730 JAC14982.1 GECL01000494 JAP05630.1 GFTR01007035 JAW09391.1 GBGD01001096 JAC87793.1 KB203566 ESO84206.1 JH432169 KK118720 KFM73686.1 GDKW01001948 JAI54647.1 CH916374 EDV91070.1 AMQN01004487 KB293638 ELU15738.1 JRES01001160 KNC25040.1 KQ424993 KOF70180.1 OUUW01000005 SPP80801.1 CM000070 EAL27010.2 AERX01017131 RQTK01000083 RUS88413.1 CH933806 EDW16523.1 KRG02179.1 GFAA01003120 JAU00315.1 CH940650 EDW67705.1 GFWV01010543 MAA35272.1 CR396584 BC125950 AAI25951.1 KQ042670 KKF12078.1 GDHF01032253 GDHF01029832 GDHF01023829 GDHF01021543 GDHF01012690 GDHF01010437 JAI20061.1 JAI22482.1 JAI28485.1 JAI30771.1 JAI39624.1 JAI41877.1 GCES01091339 JAQ94983.1 CH479186 EDW38885.1 HADW01006505 HADX01009383 SBP07905.1 CH954182 EDV53320.1 NEDP02002059 OWF51817.1 AYCK01018557 AYCK01018558 GGLE01004024 MBY08150.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000235965 UP000007755 UP000075809 UP000215335 UP000279307 UP000053097 UP000000311 UP000008237 UP000076502 UP000005205 UP000078492 UP000036403 UP000053825 UP000242457 UP000005203 UP000078540 UP000078542 UP000007266 UP000007819 UP000019118 UP000030742 UP000009046 UP000005408 UP000030746 UP000076420 UP000054359 UP000095300 UP000095301 UP000001070 UP000014760 UP000037069 UP000053454 UP000264800 UP000257200 UP000265080 UP000268350 UP000001819 UP000261460 UP000005207 UP000271974 UP000009192 UP000265140 UP000192220 UP000261620 UP000008792 UP000265100 UP000265160 UP000261340 UP000000437 UP000005226 UP000002852 UP000261400 UP000242638 UP000008744 UP000265040 UP000265020 UP000261660 UP000085678 UP000008711 UP000242188 UP000087266 UP000261420 UP000028760 UP000192221 UP000261560
UP000235965 UP000007755 UP000075809 UP000215335 UP000279307 UP000053097 UP000000311 UP000008237 UP000076502 UP000005205 UP000078492 UP000036403 UP000053825 UP000242457 UP000005203 UP000078540 UP000078542 UP000007266 UP000007819 UP000019118 UP000030742 UP000009046 UP000005408 UP000030746 UP000076420 UP000054359 UP000095300 UP000095301 UP000001070 UP000014760 UP000037069 UP000053454 UP000264800 UP000257200 UP000265080 UP000268350 UP000001819 UP000261460 UP000005207 UP000271974 UP000009192 UP000265140 UP000192220 UP000261620 UP000008792 UP000265100 UP000265160 UP000261340 UP000000437 UP000005226 UP000002852 UP000261400 UP000242638 UP000008744 UP000265040 UP000265020 UP000261660 UP000085678 UP000008711 UP000242188 UP000087266 UP000261420 UP000028760 UP000192221 UP000261560
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IYC7
A0A2W1BHN7
A0A2A4J411
A0A0N1PG23
A0A0N1PFL8
A0A212FE71
+ More
A0A067R148 A0A2J7R583 F4X8L9 A0A151WF79 A0A232F201 A0A3L8DVW3 A0A026W6T0 E2A1I7 E2BIG5 A0A154P745 A0A158NBK3 A0A195DRE1 A0A0C9PVL5 A0A0J7LA42 A0A0L7QRM3 A0A2A3ETN7 A0A088AE66 A0A195B9R9 A0A1B6G7P0 E9J9G1 A0A1B6L9E9 A0A195C6G4 A0A158NC14 A0A0C9RB08 D6W6Q1 J9JV81 A0A2H8TL98 A0A2S2P6I4 A0A2S2Q0V7 N6U5G4 A0A2J7R590 A0A0A9Z135 A0A1Y1N9I2 E0VWS5 K1QEZ2 A0A023F0I4 A0A0V0GCB7 A0A224XMZ4 A0A069DUJ6 V3ZTG6 T1JFI6 A0A2C9JQK1 A0A087U8J7 A0A1I8NN21 A0A1I8M679 A0A0P4VNU8 B4JUA0 R7VA97 A0A0L0BYE2 A0A0L8G0S4 A0A3Q3BR28 A0A3Q1FR27 A0A3P8RKM3 A0A3B0K724 Q29BI8 A0A3B4GSI6 I3JVM2 A0A3S1BPI5 B4K8E1 A0A3P8YAG8 A0A2I4DBK6 A0A3Q3VPF0 A0A2I4DBL8 A0A1E1XM36 B4LWP0 A0A3P8PI24 A0A3P9CHS7 A0A3Q0R6R2 A0A293M2Y3 A0JMN9 H2TJU3 A0A0F8AES9 A0A0K8U0J3 A0A146TQ48 A0A3B5Q704 A0A3B4ZYT3 A0A3P9MWE8 B4GP02 A0A3Q1IR09 A0A3Q2FRI4 A0A3Q3E058 A0A1S3K0H4 A0A1A7WQA9 B3P5S0 A0A210QSS9 A0A1S3MK28 A0A3B4VIU8 A0A087XLF2 A0A2I4DBJ8 A0A1W4W2J0 A0A2R5LF48 A0A3B3DY19
A0A067R148 A0A2J7R583 F4X8L9 A0A151WF79 A0A232F201 A0A3L8DVW3 A0A026W6T0 E2A1I7 E2BIG5 A0A154P745 A0A158NBK3 A0A195DRE1 A0A0C9PVL5 A0A0J7LA42 A0A0L7QRM3 A0A2A3ETN7 A0A088AE66 A0A195B9R9 A0A1B6G7P0 E9J9G1 A0A1B6L9E9 A0A195C6G4 A0A158NC14 A0A0C9RB08 D6W6Q1 J9JV81 A0A2H8TL98 A0A2S2P6I4 A0A2S2Q0V7 N6U5G4 A0A2J7R590 A0A0A9Z135 A0A1Y1N9I2 E0VWS5 K1QEZ2 A0A023F0I4 A0A0V0GCB7 A0A224XMZ4 A0A069DUJ6 V3ZTG6 T1JFI6 A0A2C9JQK1 A0A087U8J7 A0A1I8NN21 A0A1I8M679 A0A0P4VNU8 B4JUA0 R7VA97 A0A0L0BYE2 A0A0L8G0S4 A0A3Q3BR28 A0A3Q1FR27 A0A3P8RKM3 A0A3B0K724 Q29BI8 A0A3B4GSI6 I3JVM2 A0A3S1BPI5 B4K8E1 A0A3P8YAG8 A0A2I4DBK6 A0A3Q3VPF0 A0A2I4DBL8 A0A1E1XM36 B4LWP0 A0A3P8PI24 A0A3P9CHS7 A0A3Q0R6R2 A0A293M2Y3 A0JMN9 H2TJU3 A0A0F8AES9 A0A0K8U0J3 A0A146TQ48 A0A3B5Q704 A0A3B4ZYT3 A0A3P9MWE8 B4GP02 A0A3Q1IR09 A0A3Q2FRI4 A0A3Q3E058 A0A1S3K0H4 A0A1A7WQA9 B3P5S0 A0A210QSS9 A0A1S3MK28 A0A3B4VIU8 A0A087XLF2 A0A2I4DBJ8 A0A1W4W2J0 A0A2R5LF48 A0A3B3DY19
Ontologies
GO
PANTHER
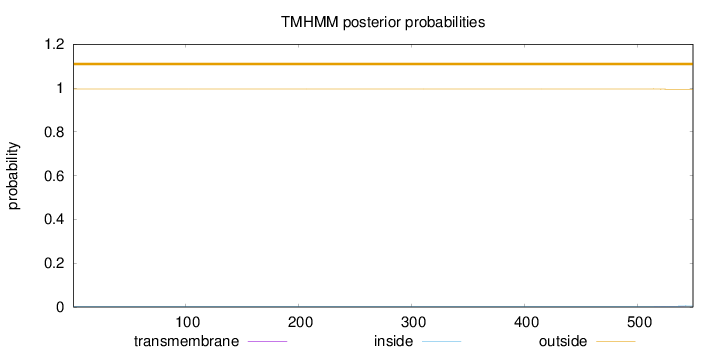
Topology
Length:
549
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02498
Exp number, first 60 AAs:
0.00089
Total prob of N-in:
0.00397
outside
1 - 549
Population Genetic Test Statistics
Pi
280.888796
Theta
159.092802
Tajima's D
2.521455
CLR
0
CSRT
0.945702714864257
Interpretation
Uncertain
