Gene
KWMTBOMO15714 Validated by peptides from experiments
Annotation
PREDICTED:_uncharacterized_protein_LOC105381048_[Plutella_xylostella]
Full name
Proliferation marker protein Ki-67
Alternative Name
Antigen identified by monoclonal antibody Ki-67
Location in the cell
Cytoplasmic Reliability : 1.056 Extracellular Reliability : 1.412 Mitochondrial Reliability : 1.102
Sequence
CDS
ATGTCAGGAGGCTGTCTGGTGGTTCTTGACCGTAATGGTCGTTATGTCAAAAAGTACCCGTTGCCTGATGGATTGGCTACATTGGGTAGTGACCCTGCATGTGATATCCGTGTGATGCTACCAACTGTCTGTTCTCATCATGCAACTGTAGTTGTGCATGCTAATCAGACTGTTGTACGAAATATTGGCGATGGCGATACCTTAGTGAATGGAGAGCCTGTGAGTGTGGCAGCGCTTAAGCACTATGATGTGATTACTTTAGGTGATCGATCTCTACGCTGGCATTATGACAATCCAAATATCAGTAAAAAATTCCAGAAGATTGAACAAGGTATGATATTGTTGATTATGAAATTCTTAAAAAACATTATAAGCAAGTACAGTTTTCCTTAA
Protein
MSGGCLVVLDRNGRYVKKYPLPDGLATLGSDPACDIRVMLPTVCSHHATVVVHANQTVVRNIGDGDTLVNGEPVSVAALKHYDVITLGDRSLRWHYDNPNISKKFQKIEQGMILLIMKFLKNIISKYSFP
Summary
Description
Required to maintain individual mitotic chromosomes dispersed in the cytoplasm following nuclear envelope disassembly (PubMed:27362226). Associates with the surface of the mitotic chromosome, the perichromosomal layer, and covers a substantial fraction of the chromosome surface (PubMed:27362226). Prevents chromosomes from collapsing into a single chromatin mass by forming a steric and electrostatic charge barrier: the protein has a high net electrical charge and acts as a surfactant, dispersing chromosomes and enabling independent chromosome motility (PubMed:27362226). Binds DNA, with a preference for supercoiled DNA and AT-rich DNA (PubMed:10878551). Does not contribute to the internal structure of mitotic chromosomes (By similarity). May play a role in chromatin organization (PubMed:24867636). It is however unclear whether it plays a direct role in chromatin organization or whether it is an indirect consequence of its function in maintaining mitotic chromosomes dispersed (Probable).
Subunit
Interacts with KIF15 (PubMed:10878014). Interacts (via the FHA domain) with NIFK (PubMed:11342549, PubMed:14659764, PubMed:16244663). Interacts with PPP1CC (PubMed:24867636). Part of a complex composed at least of ASCL2, EMSY, HCFC1, HSPA8, CCAR2, MATR3, MKI67, RBBP5, TUBB2A, WDR5 and ZNF335; this complex may have a histone H3-specific methyltransferase activity (PubMed:19131338, PubMed:23178126).
Keywords
3D-structure
Acetylation
Alternative splicing
ATP-binding
Cell cycle
Chromosome
Complete proteome
DNA-binding
Isopeptide bond
Nucleotide-binding
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Repeat
Ubl conjugation
Feature
chain Proliferation marker protein Ki-67
splice variant In isoform Short.
sequence variant In dbSNP:rs2071498.
splice variant In isoform Short.
sequence variant In dbSNP:rs2071498.
Uniprot
A0A2W1BHQ3
A0A0N0PBS5
A0A0N1IN47
A0A212FE84
A0A2B4RY07
A0A1B6H0L0
+ More
A0A1B6EWL0 A0A093GR25 A0A1B6HL51 A0A1B6L3C4 A7SHF7 A0A1B6G5C1 A0A1B6GV61 A0A3P8PRT5 A0A3Q2W592 F6VDD5 A0A2D4N0H4 A0A2D4N2A9 A0A0S7HQ39 A0A3N1YBA2 A0A091Q5K5 G1SNA0 A0A091GU99 A0A091SWM9 V3ZNX7 A0A091G6S3 H2UYK8 A0A3Q3LFX4 A0A3B5KCG6 B5XEL4 A0A093EPU6 A0A087QZR1 A0A1W2WNZ4 A0A093HWS1 A0A1S2X433 A0A2I4B743 A0A2I4B756 A0A091JLT9 A0A2I4B763 A0A091LBI5 A0A091TG21 A0A093NUD3 A0A091S3L4 A0A091UNR2 A0A087V6V0 I3KVD4 A0A091HVT4 A0A094L8T5 A0A093QBA2 A0A2U9CNQ4 A0A0A0AZQ1 A0A091M352 R7VRK2 A0A099Z8Q1 T2HWM1 H2YXT6 H3D5N7 G1NGY9 A0A2K5RSS1 I3KVD5 A0A315W966 A0A2K5RSQ0 A0A3B3XNF4 A0A3P9Q827 A0A3B3UXP5 A0A087YE19 A0A096M7S8 A0A3Q3AA26 A0A091KWK0 A0A2K6TDP6 A0A2K6TDP7 A0A3Q3FJ07 A0A3B4YLF4 P46013-2 Q4S4N2 P46013 A0A3B4TFH4 A0A093HHV6 A0A3P9JM20 A0A3P9ICG3 A0A3B3IBR9 A0A3P9LGX2 A0A091VGD0 A0A094KNR2 A0A3Q1BRE4 M4AJK8 W7TDZ3 A0A3Q1F9F8 K7FV23 A0A3Q2L9G7 A0A2I3GRS6 A0A2D0S6G5 G1QLE5 F7AFX1 A0A3Q3DIN8 A0A3B5BCL1 F6VGN1
A0A1B6EWL0 A0A093GR25 A0A1B6HL51 A0A1B6L3C4 A7SHF7 A0A1B6G5C1 A0A1B6GV61 A0A3P8PRT5 A0A3Q2W592 F6VDD5 A0A2D4N0H4 A0A2D4N2A9 A0A0S7HQ39 A0A3N1YBA2 A0A091Q5K5 G1SNA0 A0A091GU99 A0A091SWM9 V3ZNX7 A0A091G6S3 H2UYK8 A0A3Q3LFX4 A0A3B5KCG6 B5XEL4 A0A093EPU6 A0A087QZR1 A0A1W2WNZ4 A0A093HWS1 A0A1S2X433 A0A2I4B743 A0A2I4B756 A0A091JLT9 A0A2I4B763 A0A091LBI5 A0A091TG21 A0A093NUD3 A0A091S3L4 A0A091UNR2 A0A087V6V0 I3KVD4 A0A091HVT4 A0A094L8T5 A0A093QBA2 A0A2U9CNQ4 A0A0A0AZQ1 A0A091M352 R7VRK2 A0A099Z8Q1 T2HWM1 H2YXT6 H3D5N7 G1NGY9 A0A2K5RSS1 I3KVD5 A0A315W966 A0A2K5RSQ0 A0A3B3XNF4 A0A3P9Q827 A0A3B3UXP5 A0A087YE19 A0A096M7S8 A0A3Q3AA26 A0A091KWK0 A0A2K6TDP6 A0A2K6TDP7 A0A3Q3FJ07 A0A3B4YLF4 P46013-2 Q4S4N2 P46013 A0A3B4TFH4 A0A093HHV6 A0A3P9JM20 A0A3P9ICG3 A0A3B3IBR9 A0A3P9LGX2 A0A091VGD0 A0A094KNR2 A0A3Q1BRE4 M4AJK8 W7TDZ3 A0A3Q1F9F8 K7FV23 A0A3Q2L9G7 A0A2I3GRS6 A0A2D0S6G5 G1QLE5 F7AFX1 A0A3Q3DIN8 A0A3B5BCL1 F6VGN1
Pubmed
28756777
26354079
22118469
17615350
12481130
15114417
+ More
21993624 23254933 21551351 20433749 25186727 23371554 24019933 15496914 20838655 29703783 8227122 15164054 6339421 6206131 2674163 8799815 9510506 10502411 10878014 10653604 10878551 11342549 15896774 17081983 16964243 17924679 18220336 18691976 18669648 19413330 19131338 19369195 19690332 19608861 20068231 21960707 21406692 23178126 22002106 23186163 24867636 25218447 25114211 25772364 25755297 26680267 27362226 28112733 14659764 16244663 17554307 23542700 23966634 17381049 19892987 20431018
21993624 23254933 21551351 20433749 25186727 23371554 24019933 15496914 20838655 29703783 8227122 15164054 6339421 6206131 2674163 8799815 9510506 10502411 10878014 10653604 10878551 11342549 15896774 17081983 16964243 17924679 18220336 18691976 18669648 19413330 19131338 19369195 19690332 19608861 20068231 21960707 21406692 23178126 22002106 23186163 24867636 25218447 25114211 25772364 25755297 26680267 27362226 28112733 14659764 16244663 17554307 23542700 23966634 17381049 19892987 20431018
EMBL
KZ150119
PZC73224.1
KQ460820
KPJ12109.1
KQ459199
KPJ02969.1
+ More
AGBW02008963 OWR52066.1 LSMT01000290 PFX21132.1 GECZ01001553 JAS68216.1 GECZ01027395 JAS42374.1 KL217011 KFV72758.1 GECU01032300 JAS75406.1 GEBQ01021780 JAT18197.1 DS469659 EDO36884.1 GECZ01012131 JAS57638.1 GECZ01003463 JAS66306.1 EAAA01001584 IACM01135423 LAB39155.1 IACM01135424 LAB39156.1 GBYX01438309 JAO43044.1 RJVI01000001 ROR34677.1 KK687464 KFQ15185.1 KL513362 KFO87191.1 KK489663 KFQ63084.1 KB201977 ESO93083.1 KL447873 KFO78090.1 BT049483 ACI69284.2 KK366919 KFV41544.1 KL226002 KFM06715.1 KK636914 KFV58953.1 KK501100 KFP12666.1 KL308756 KFP52673.1 KK449719 KFQ72825.1 KL225167 KFW68393.1 KK939502 KFQ49578.1 KL409816 KFQ92012.1 KL482157 KFO08342.1 AERX01002294 AERX01002295 KL217818 KFO99269.1 KL281907 KFZ67728.1 KL418178 KFW85876.1 CP026260 AWP17479.1 KL873581 KGL99417.1 KK516965 KFP65074.1 KB375595 EMC84126.1 KL891498 KGL78864.1 AB775130 BAN83908.1 NHOQ01000843 PWA28382.1 AYCK01000749 AYCK01000750 KK544981 KFP32064.1 X65550 X65551 AL355529 AL390236 X94762 CAAE01014738 CAG04400.1 KL206121 KFV79030.1 KK733934 KFR01508.1 KL353206 KFZ60260.1 AZIL01001014 EWM25215.1 AGCU01074131 ADFV01087722 AAMC01014933 AAMC01014934
AGBW02008963 OWR52066.1 LSMT01000290 PFX21132.1 GECZ01001553 JAS68216.1 GECZ01027395 JAS42374.1 KL217011 KFV72758.1 GECU01032300 JAS75406.1 GEBQ01021780 JAT18197.1 DS469659 EDO36884.1 GECZ01012131 JAS57638.1 GECZ01003463 JAS66306.1 EAAA01001584 IACM01135423 LAB39155.1 IACM01135424 LAB39156.1 GBYX01438309 JAO43044.1 RJVI01000001 ROR34677.1 KK687464 KFQ15185.1 KL513362 KFO87191.1 KK489663 KFQ63084.1 KB201977 ESO93083.1 KL447873 KFO78090.1 BT049483 ACI69284.2 KK366919 KFV41544.1 KL226002 KFM06715.1 KK636914 KFV58953.1 KK501100 KFP12666.1 KL308756 KFP52673.1 KK449719 KFQ72825.1 KL225167 KFW68393.1 KK939502 KFQ49578.1 KL409816 KFQ92012.1 KL482157 KFO08342.1 AERX01002294 AERX01002295 KL217818 KFO99269.1 KL281907 KFZ67728.1 KL418178 KFW85876.1 CP026260 AWP17479.1 KL873581 KGL99417.1 KK516965 KFP65074.1 KB375595 EMC84126.1 KL891498 KGL78864.1 AB775130 BAN83908.1 NHOQ01000843 PWA28382.1 AYCK01000749 AYCK01000750 KK544981 KFP32064.1 X65550 X65551 AL355529 AL390236 X94762 CAAE01014738 CAG04400.1 KL206121 KFV79030.1 KK733934 KFR01508.1 KL353206 KFZ60260.1 AZIL01001014 EWM25215.1 AGCU01074131 ADFV01087722 AAMC01014933 AAMC01014934
Proteomes
UP000053240
UP000053268
UP000007151
UP000225706
UP000053875
UP000001593
+ More
UP000265100 UP000264840 UP000008144 UP000276634 UP000001811 UP000030746 UP000053760 UP000005226 UP000261640 UP000053286 UP000087266 UP000192220 UP000053119 UP000054081 UP000053283 UP000005207 UP000054308 UP000246464 UP000053858 UP000053641 UP000007875 UP000007303 UP000001645 UP000233040 UP000261480 UP000242638 UP000261500 UP000028760 UP000264800 UP000233220 UP000261660 UP000261360 UP000005640 UP000261420 UP000053584 UP000265200 UP000001038 UP000265180 UP000053605 UP000257160 UP000002852 UP000257200 UP000007267 UP000002281 UP000001073 UP000221080 UP000008143 UP000264820 UP000261400
UP000265100 UP000264840 UP000008144 UP000276634 UP000001811 UP000030746 UP000053760 UP000005226 UP000261640 UP000053286 UP000087266 UP000192220 UP000053119 UP000054081 UP000053283 UP000005207 UP000054308 UP000246464 UP000053858 UP000053641 UP000007875 UP000007303 UP000001645 UP000233040 UP000261480 UP000242638 UP000261500 UP000028760 UP000264800 UP000233220 UP000261660 UP000261360 UP000005640 UP000261420 UP000053584 UP000265200 UP000001038 UP000265180 UP000053605 UP000257160 UP000002852 UP000257200 UP000007267 UP000002281 UP000001073 UP000221080 UP000008143 UP000264820 UP000261400
PRIDE
Interpro
SUPFAM
SSF49879
SSF49879
CDD
ProteinModelPortal
A0A2W1BHQ3
A0A0N0PBS5
A0A0N1IN47
A0A212FE84
A0A2B4RY07
A0A1B6H0L0
+ More
A0A1B6EWL0 A0A093GR25 A0A1B6HL51 A0A1B6L3C4 A7SHF7 A0A1B6G5C1 A0A1B6GV61 A0A3P8PRT5 A0A3Q2W592 F6VDD5 A0A2D4N0H4 A0A2D4N2A9 A0A0S7HQ39 A0A3N1YBA2 A0A091Q5K5 G1SNA0 A0A091GU99 A0A091SWM9 V3ZNX7 A0A091G6S3 H2UYK8 A0A3Q3LFX4 A0A3B5KCG6 B5XEL4 A0A093EPU6 A0A087QZR1 A0A1W2WNZ4 A0A093HWS1 A0A1S2X433 A0A2I4B743 A0A2I4B756 A0A091JLT9 A0A2I4B763 A0A091LBI5 A0A091TG21 A0A093NUD3 A0A091S3L4 A0A091UNR2 A0A087V6V0 I3KVD4 A0A091HVT4 A0A094L8T5 A0A093QBA2 A0A2U9CNQ4 A0A0A0AZQ1 A0A091M352 R7VRK2 A0A099Z8Q1 T2HWM1 H2YXT6 H3D5N7 G1NGY9 A0A2K5RSS1 I3KVD5 A0A315W966 A0A2K5RSQ0 A0A3B3XNF4 A0A3P9Q827 A0A3B3UXP5 A0A087YE19 A0A096M7S8 A0A3Q3AA26 A0A091KWK0 A0A2K6TDP6 A0A2K6TDP7 A0A3Q3FJ07 A0A3B4YLF4 P46013-2 Q4S4N2 P46013 A0A3B4TFH4 A0A093HHV6 A0A3P9JM20 A0A3P9ICG3 A0A3B3IBR9 A0A3P9LGX2 A0A091VGD0 A0A094KNR2 A0A3Q1BRE4 M4AJK8 W7TDZ3 A0A3Q1F9F8 K7FV23 A0A3Q2L9G7 A0A2I3GRS6 A0A2D0S6G5 G1QLE5 F7AFX1 A0A3Q3DIN8 A0A3B5BCL1 F6VGN1
A0A1B6EWL0 A0A093GR25 A0A1B6HL51 A0A1B6L3C4 A7SHF7 A0A1B6G5C1 A0A1B6GV61 A0A3P8PRT5 A0A3Q2W592 F6VDD5 A0A2D4N0H4 A0A2D4N2A9 A0A0S7HQ39 A0A3N1YBA2 A0A091Q5K5 G1SNA0 A0A091GU99 A0A091SWM9 V3ZNX7 A0A091G6S3 H2UYK8 A0A3Q3LFX4 A0A3B5KCG6 B5XEL4 A0A093EPU6 A0A087QZR1 A0A1W2WNZ4 A0A093HWS1 A0A1S2X433 A0A2I4B743 A0A2I4B756 A0A091JLT9 A0A2I4B763 A0A091LBI5 A0A091TG21 A0A093NUD3 A0A091S3L4 A0A091UNR2 A0A087V6V0 I3KVD4 A0A091HVT4 A0A094L8T5 A0A093QBA2 A0A2U9CNQ4 A0A0A0AZQ1 A0A091M352 R7VRK2 A0A099Z8Q1 T2HWM1 H2YXT6 H3D5N7 G1NGY9 A0A2K5RSS1 I3KVD5 A0A315W966 A0A2K5RSQ0 A0A3B3XNF4 A0A3P9Q827 A0A3B3UXP5 A0A087YE19 A0A096M7S8 A0A3Q3AA26 A0A091KWK0 A0A2K6TDP6 A0A2K6TDP7 A0A3Q3FJ07 A0A3B4YLF4 P46013-2 Q4S4N2 P46013 A0A3B4TFH4 A0A093HHV6 A0A3P9JM20 A0A3P9ICG3 A0A3B3IBR9 A0A3P9LGX2 A0A091VGD0 A0A094KNR2 A0A3Q1BRE4 M4AJK8 W7TDZ3 A0A3Q1F9F8 K7FV23 A0A3Q2L9G7 A0A2I3GRS6 A0A2D0S6G5 G1QLE5 F7AFX1 A0A3Q3DIN8 A0A3B5BCL1 F6VGN1
PDB
1R21
E-value=3.23365e-09,
Score=140
Ontologies
GO
Topology
Subcellular location
Chromosome
Associates with the surface of the mitotic chromosome, the perichromosomal layer, and covers a substantial fraction of the mitotic chromosome surface (PubMed:27362226). Associates with satellite DNA in G1 phase (PubMed:9510506). Binds tightly to chromatin in interphase, chromatin-binding decreases in mitosis when it associates with the surface of the condensed chromosomes (PubMed:15896774, PubMed:22002106). Predominantly localized in the G1 phase in the perinucleolar region, in the later phases it is also detected throughout the nuclear interior, being predominantly localized in the nuclear matrix (PubMed:22002106). With evidence from 30 publications.
Nucleus Associates with the surface of the mitotic chromosome, the perichromosomal layer, and covers a substantial fraction of the mitotic chromosome surface (PubMed:27362226). Associates with satellite DNA in G1 phase (PubMed:9510506). Binds tightly to chromatin in interphase, chromatin-binding decreases in mitosis when it associates with the surface of the condensed chromosomes (PubMed:15896774, PubMed:22002106). Predominantly localized in the G1 phase in the perinucleolar region, in the later phases it is also detected throughout the nuclear interior, being predominantly localized in the nuclear matrix (PubMed:22002106). With evidence from 30 publications.
Nucleolus Associates with the surface of the mitotic chromosome, the perichromosomal layer, and covers a substantial fraction of the mitotic chromosome surface (PubMed:27362226). Associates with satellite DNA in G1 phase (PubMed:9510506). Binds tightly to chromatin in interphase, chromatin-binding decreases in mitosis when it associates with the surface of the condensed chromosomes (PubMed:15896774, PubMed:22002106). Predominantly localized in the G1 phase in the perinucleolar region, in the later phases it is also detected throughout the nuclear interior, being predominantly localized in the nuclear matrix (PubMed:22002106). With evidence from 30 publications.
Nucleus Associates with the surface of the mitotic chromosome, the perichromosomal layer, and covers a substantial fraction of the mitotic chromosome surface (PubMed:27362226). Associates with satellite DNA in G1 phase (PubMed:9510506). Binds tightly to chromatin in interphase, chromatin-binding decreases in mitosis when it associates with the surface of the condensed chromosomes (PubMed:15896774, PubMed:22002106). Predominantly localized in the G1 phase in the perinucleolar region, in the later phases it is also detected throughout the nuclear interior, being predominantly localized in the nuclear matrix (PubMed:22002106). With evidence from 30 publications.
Nucleolus Associates with the surface of the mitotic chromosome, the perichromosomal layer, and covers a substantial fraction of the mitotic chromosome surface (PubMed:27362226). Associates with satellite DNA in G1 phase (PubMed:9510506). Binds tightly to chromatin in interphase, chromatin-binding decreases in mitosis when it associates with the surface of the condensed chromosomes (PubMed:15896774, PubMed:22002106). Predominantly localized in the G1 phase in the perinucleolar region, in the later phases it is also detected throughout the nuclear interior, being predominantly localized in the nuclear matrix (PubMed:22002106). With evidence from 30 publications.
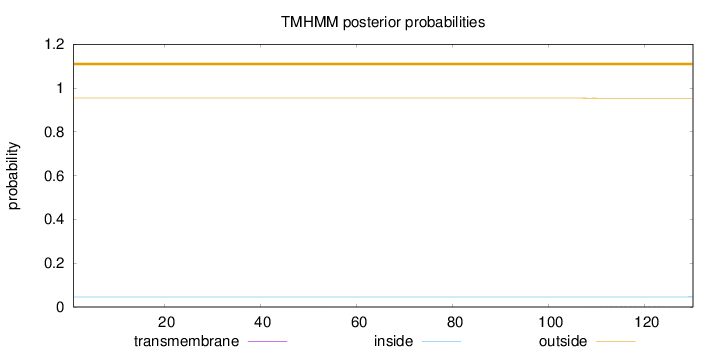
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02773
Exp number, first 60 AAs:
0.00423
Total prob of N-in:
0.04551
outside
1 - 130
Population Genetic Test Statistics
Pi
213.460627
Theta
184.434231
Tajima's D
-0.793491
CLR
0
CSRT
0.179491025448728
Interpretation
Uncertain
