Gene
KWMTBOMO15712 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002309
Annotation
nucleoplasmin_isoform_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.936
Sequence
CDS
ATGACGGACGAGTTTTTCTATGGTGTCACCCTTTCATCATCACATCAGTCAGAGACATGGGATCCAGAGGCAAAAGCAGAATACCCACGCAGCAACAAGCTCGTCATTCGTCAAGCATTGTTAGGTCCAGATGCCAAACCAGATGAATTAAATGTGATACAGGTGGAAGCCATGTCACTACAAGAAGCAGTAAAACTACCAGTCGCAGTATTGAAAGTTGGTGAATCAAGGCATGTACGTCTGGACATTGAATTCCCAGATGCGCCTGTTACATTTACTCTTGTTCAGGGTTCGGGGCCAGTACACTTAATTGGACACCATCTCCTTGGTGCTCTGCTTGAAGAATTTGAAGATATGGAGGAAATGGAAGAAGAAATGTTAGATGAAGAAGAAGGAGATGATTCTCAGTTCAAGGAAGATGAGAATAAAAGAAAAGGAGCAGGGAAGCGCAAGCCGAATGAGGATGAAGATAATGAAGAAGGTGAACCCAAGGGCAAAAAAGCGAAAATGTCGAATAATGCCAAAGGCAAAGCTGCATCGCCCAAGAAGAATGCCAAGAAATGA
Protein
MTDEFFYGVTLSSSHQSETWDPEAKAEYPRSNKLVIRQALLGPDAKPDELNVIQVEAMSLQEAVKLPVAVLKVGESRHVRLDIEFPDAPVTFTLVQGSGPVHLIGHHLLGALLEEFEDMEEMEEEMLDEEEGDDSQFKEDENKRKGAGKRKPNEDEDNEEGEPKGKKAKMSNNAKGKAASPKKNAKK
Summary
Uniprot
EMBL
BABH01020634
DQ311384
ABD36328.1
KQ460820
KPJ12111.1
AK401336
+ More
KQ459199 BAM17958.1 KPJ02971.1 KZ150119 PZC73222.1 ODYU01000822 SOQ36177.1 NWSH01006279 PCG63534.1 DQ311383 ABD36327.1 RSAL01000022 RVE52323.1 AGBW02008963 OWR52068.1 EF210318 ABN09649.1 GDQN01005644 JAT85410.1 AK402602 BAM19224.1 PCG63532.1 GDQN01004540 JAT86514.1
KQ459199 BAM17958.1 KPJ02971.1 KZ150119 PZC73222.1 ODYU01000822 SOQ36177.1 NWSH01006279 PCG63534.1 DQ311383 ABD36327.1 RSAL01000022 RVE52323.1 AGBW02008963 OWR52068.1 EF210318 ABN09649.1 GDQN01005644 JAT85410.1 AK402602 BAM19224.1 PCG63532.1 GDQN01004540 JAT86514.1
Proteomes
Pfam
PF03066 Nucleoplasmin
Interpro
SUPFAM
SSF69203
SSF69203
ProteinModelPortal
PDB
1NLQ
E-value=1.14434e-19,
Score=233
Ontologies
GO
PANTHER
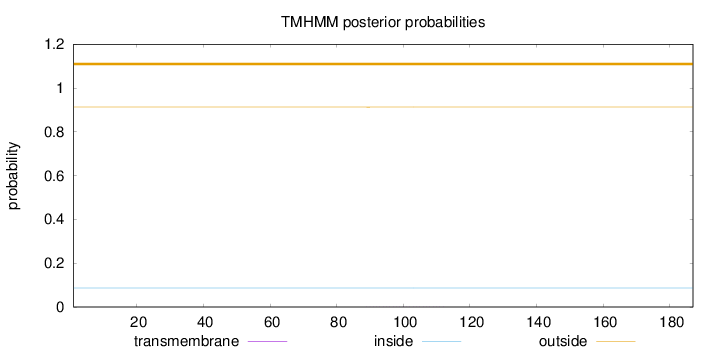
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00882
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.08674
outside
1 - 187
Population Genetic Test Statistics
Pi
197.297687
Theta
163.181283
Tajima's D
0.972216
CLR
0
CSRT
0.650517474126294
Interpretation
Uncertain
