Gene
KWMTBOMO15710 Validated by peptides from experiments
Annotation
PREDICTED:_actin-binding_LIM_protein_1-like_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 3.778
Sequence
CDS
ATGAATGGCAAAGTGGTGTGCGCATCTTGCGGCGGCAAGTGCAGCGGTGAGGTGCTTCGCGTGTCGGACAAGTACTTCCACACCGCCTGCTTCACGTGCCGCACTTGTTCAGCCTCTCTCGCCAAAGGGGGCTTCTTCTGCAAGGACGGACACTACTACTGCCCGCAGGATTACCAACGCGCGTTTGGCACACGATGCGCAGCCTGCAACCAGTATGTAGAAGGAGAGGTGGTCTCCGCCCTCGGGAATACGTATCATCAGAAATGCTTCACTTGTGCGAGATGCAAGAGAGCGTTCCCGTCGGGTGAGAAAGTGACGTATACAGGAACAGAAGTTGTCTGTTCGAACTGCGTGGCGGTGCCGCAGCGACAGACCGCGAGGACTGCGGCGCCGGCCCCCCTCAGCTCCCCCACTCCGGAGCCACAGCAGAATAATGTTGAACGACGACAGCCAGATCAGAACGAATGCGCTGGTTGCGGTCAAGAATTATCCGAGGGTCAGGCTTTGGTGGCACTAGACAGACAATGGCACACTGGGTGCTTCTCGTGCGGCGAGTGCGGCGCCGTGCTGCAGGGCGAGTACATGGGCCGGGACGGCGTGCCTTACTGCGAGCGAGACTACCAGCGTCTGTACGGAGTGCGGTGCGCGTACTGCCAGCGATACATATCCGGGAAGGTGTTGCAGGCCGGAGAGAATCACCACTTCCACCCGACGTGCGCGCGCTGCAGCAAGTGCGGAGATCCATTCGGCGACGGCGAGGAGATGTTCCTCCAGGGCGCCGCCATCTGGCACCCGCGCTGCGGGCCCGCGCCCCACGACCCGCTCTCCGCCGATAGAGCCTCCTCCGAGTTGCAGTTCTCGCTGCGATCGCGCACACCCAGCGTCAACGGCTCATACTGCAGTCCCTACAGTAGCTTGACTAGAAAGTACGGGTACCGAGCCGTGTCCCCCGGGCTGGTGCTACGCGAGTACCGCGAGGCGGGCGCGTGCAGTCCGCACCGCATCACCACGTACTCGTACCTCACGACGGAGCCTGCGTACCTGCGCCGCCCCGTGCAGCCCTACGACCGCCCGCCCACCTCGCCGCACTTCCACCGCCCGCCCTGTAAGTAA
Protein
MNGKVVCASCGGKCSGEVLRVSDKYFHTACFTCRTCSASLAKGGFFCKDGHYYCPQDYQRAFGTRCAACNQYVEGEVVSALGNTYHQKCFTCARCKRAFPSGEKVTYTGTEVVCSNCVAVPQRQTARTAAPAPLSSPTPEPQQNNVERRQPDQNECAGCGQELSEGQALVALDRQWHTGCFSCGECGAVLQGEYMGRDGVPYCERDYQRLYGVRCAYCQRYISGKVLQAGENHHFHPTCARCSKCGDPFGDGEEMFLQGAAIWHPRCGPAPHDPLSADRASSELQFSLRSRTPSVNGSYCSPYSSLTRKYGYRAVSPGLVLREYREAGACSPHRITTYSYLTTEPAYLRRPVQPYDRPPTSPHFHRPPCK
Summary
Uniprot
A0A2W1BDB1
A0A2H1W9Y7
A0A0N1IEY7
A0A212FEB5
V5GP13
A0A1W4W4N6
+ More
A0A1W4WGH3 A0A1W4WF73 A0A1W4WG72 A0A1W4W4M6 A0A2J7PJD5 A0A1W4WGG7 A0A2J7PJE1 A0A1Y1K033 Q7Q796 A0A1Y1JYF0 A0A1Y1JSP2 A0A182LTL8 D6X362 R4WQ91 Q0IEQ0 A0A1Y1JXC0 T1DCM8 A0A182WLU5 A0A2M3YY63 A0A2M4A2N4 A0A2M4A2R8 A0A2M4CY33 A0A336KE08 N6UMK8 A0A182RC20 A0A182FHN8 A0A1Q3FEV7 A0A1J1J283 U5EZV4 A0A1D2N7A2 A0A1Q3FEX1 A0A084VD99 A0A1Q3FF14 W5JD97 A0A0P5XNC5 A0A0P4XUL7 A0A0P6GFV7 A0A0P5QZ90 A0A0P5AD46 A0A0P5ZZM6 A0A0P5EKG4 A0A0P5DBB7 A0A0P5D9Z7 E9FUH3 A0A0N8A0F8 A0A0P5XYF5 A0A0P5KA17 A0A0P4XND8 A0A0P5XAD4 A0A1W4W4N2 A0A0N8A7G3 A0A0P4Z4X1 A0A1W4WF78 A0A0P4XJP7 A0A0P5WBE5 A0A0P5VMZ3 A0A0N8DVI5 A0A0P5XWA6 A0A0P5XCQ4 A0A0P5LUK6 A0A0P5NMB3 A0A0P5P4M8 A0A182LJT8 A0A182H289 A0A0K8VRG5 A0A1Y1JS46 W8BSZ4 U4UGV7 A0A1I8NEN7 T1PEY8 A0A0K8V6T8 A0A0K8UUZ2 W8AU25 A0A1I8NEP4 A0A2J7PJE0 A0A034W3D2 A0A1Y1JSQ0 A0A034W2E0 A0A034W0S3 A0A0K8U3U7 E0VTH7 A0A0A1WK92 A0A1I8NEN3 A0A1I8NEP7 A0A0P4WAZ9 E2BLU7 W8B1G0 A0A158NGA8 A0A195CN49 A0A088ABY8 A0A336LYN1 A0A195E6A0 A0A023FAJ6 A0A0P5W8X5
A0A1W4WGH3 A0A1W4WF73 A0A1W4WG72 A0A1W4W4M6 A0A2J7PJD5 A0A1W4WGG7 A0A2J7PJE1 A0A1Y1K033 Q7Q796 A0A1Y1JYF0 A0A1Y1JSP2 A0A182LTL8 D6X362 R4WQ91 Q0IEQ0 A0A1Y1JXC0 T1DCM8 A0A182WLU5 A0A2M3YY63 A0A2M4A2N4 A0A2M4A2R8 A0A2M4CY33 A0A336KE08 N6UMK8 A0A182RC20 A0A182FHN8 A0A1Q3FEV7 A0A1J1J283 U5EZV4 A0A1D2N7A2 A0A1Q3FEX1 A0A084VD99 A0A1Q3FF14 W5JD97 A0A0P5XNC5 A0A0P4XUL7 A0A0P6GFV7 A0A0P5QZ90 A0A0P5AD46 A0A0P5ZZM6 A0A0P5EKG4 A0A0P5DBB7 A0A0P5D9Z7 E9FUH3 A0A0N8A0F8 A0A0P5XYF5 A0A0P5KA17 A0A0P4XND8 A0A0P5XAD4 A0A1W4W4N2 A0A0N8A7G3 A0A0P4Z4X1 A0A1W4WF78 A0A0P4XJP7 A0A0P5WBE5 A0A0P5VMZ3 A0A0N8DVI5 A0A0P5XWA6 A0A0P5XCQ4 A0A0P5LUK6 A0A0P5NMB3 A0A0P5P4M8 A0A182LJT8 A0A182H289 A0A0K8VRG5 A0A1Y1JS46 W8BSZ4 U4UGV7 A0A1I8NEN7 T1PEY8 A0A0K8V6T8 A0A0K8UUZ2 W8AU25 A0A1I8NEP4 A0A2J7PJE0 A0A034W3D2 A0A1Y1JSQ0 A0A034W2E0 A0A034W0S3 A0A0K8U3U7 E0VTH7 A0A0A1WK92 A0A1I8NEN3 A0A1I8NEP7 A0A0P4WAZ9 E2BLU7 W8B1G0 A0A158NGA8 A0A195CN49 A0A088ABY8 A0A336LYN1 A0A195E6A0 A0A023FAJ6 A0A0P5W8X5
Pubmed
EMBL
KZ150119
PZC73219.1
ODYU01007258
SOQ49878.1
KQ460820
KPJ12114.1
+ More
AGBW02008963 OWR52074.1 GALX01005124 JAB63342.1 NEVH01024949 PNF16455.1 PNF16457.1 GEZM01102039 JAV52037.1 AAAB01008960 EAA11937.4 GEZM01102040 JAV52036.1 GEZM01102042 GEZM01102037 JAV52033.1 AXCM01001428 KQ971372 EFA10334.2 AK417861 BAN21076.1 CH477504 EAT39880.1 GEZM01102043 GEZM01102041 GEZM01102038 JAV52035.1 GALA01001767 JAA93085.1 GGFM01000458 MBW21209.1 GGFK01001708 MBW35029.1 GGFK01001710 MBW35031.1 GGFL01006039 MBW70217.1 UFQS01000309 UFQT01000309 SSX02719.1 SSX23093.1 APGK01017170 APGK01017171 APGK01017172 KB739998 ENN81936.1 GFDL01008970 JAV26075.1 CVRI01000066 CRL05884.1 GANO01000074 JAB59797.1 LJIJ01000169 ODN01148.1 GFDL01008951 JAV26094.1 ATLV01011195 KE524650 KFB35943.1 GFDL01008914 JAV26131.1 ADMH02001568 ETN62046.1 GDIP01069589 JAM34126.1 GDIP01236415 JAI86986.1 GDIQ01053799 JAN40938.1 GDIQ01160963 GDIQ01113520 JAL38206.1 GDIP01200670 JAJ22732.1 GDIP01036893 JAM66822.1 GDIP01158533 JAJ64869.1 GDIP01160222 JAJ63180.1 GDIP01160780 JAJ62622.1 GL732525 EFX88923.1 GDIP01194673 JAJ28729.1 GDIP01232762 GDIP01175034 GDIP01066339 GDIQ01086099 JAM37376.1 JAN08638.1 GDIQ01188593 GDIP01141428 JAK63132.1 JAL62286.1 GDIP01239222 JAI84179.1 GDIP01074994 JAM28721.1 GDIP01175033 GDIP01141429 GDIP01036892 JAJ48369.1 GDIP01217918 JAJ05484.1 GDIP01240376 JAI83025.1 GDIP01088483 JAM15232.1 GDIP01097840 JAM05875.1 GDIQ01087788 JAN06949.1 GDIP01067828 JAM35887.1 GDIP01074996 JAM28719.1 GDIQ01164864 JAK86861.1 GDIQ01182302 GDIQ01162357 JAK89368.1 GDIP01175036 GDIQ01163515 GDIQ01133770 JAJ48366.1 JAL17956.1 JXUM01023685 JXUM01023686 JXUM01023687 JXUM01023688 KQ560668 KXJ81360.1 GDHF01010842 JAI41472.1 GEZM01102036 JAV52042.1 GAMC01014216 JAB92339.1 KB632318 ERL92262.1 KA646463 AFP61092.1 GDHF01018034 JAI34280.1 GDHF01021822 JAI30492.1 GAMC01014215 JAB92340.1 PNF16456.1 GAKP01010684 JAC48268.1 GEZM01102035 GEZM01102034 JAV52043.1 GAKP01010687 JAC48265.1 GAKP01010683 JAC48269.1 GDHF01031101 JAI21213.1 DS235766 EEB16683.1 GBXI01014818 JAC99473.1 GDRN01077549 JAI62719.1 GL449042 EFN83313.1 GAMC01014218 JAB92337.1 ADTU01015113 KQ977574 KYN01897.1 SSX02720.1 SSX23094.1 KQ979608 KYN20374.1 GBBI01000235 JAC18477.1 GDIP01090734 JAM12981.1
AGBW02008963 OWR52074.1 GALX01005124 JAB63342.1 NEVH01024949 PNF16455.1 PNF16457.1 GEZM01102039 JAV52037.1 AAAB01008960 EAA11937.4 GEZM01102040 JAV52036.1 GEZM01102042 GEZM01102037 JAV52033.1 AXCM01001428 KQ971372 EFA10334.2 AK417861 BAN21076.1 CH477504 EAT39880.1 GEZM01102043 GEZM01102041 GEZM01102038 JAV52035.1 GALA01001767 JAA93085.1 GGFM01000458 MBW21209.1 GGFK01001708 MBW35029.1 GGFK01001710 MBW35031.1 GGFL01006039 MBW70217.1 UFQS01000309 UFQT01000309 SSX02719.1 SSX23093.1 APGK01017170 APGK01017171 APGK01017172 KB739998 ENN81936.1 GFDL01008970 JAV26075.1 CVRI01000066 CRL05884.1 GANO01000074 JAB59797.1 LJIJ01000169 ODN01148.1 GFDL01008951 JAV26094.1 ATLV01011195 KE524650 KFB35943.1 GFDL01008914 JAV26131.1 ADMH02001568 ETN62046.1 GDIP01069589 JAM34126.1 GDIP01236415 JAI86986.1 GDIQ01053799 JAN40938.1 GDIQ01160963 GDIQ01113520 JAL38206.1 GDIP01200670 JAJ22732.1 GDIP01036893 JAM66822.1 GDIP01158533 JAJ64869.1 GDIP01160222 JAJ63180.1 GDIP01160780 JAJ62622.1 GL732525 EFX88923.1 GDIP01194673 JAJ28729.1 GDIP01232762 GDIP01175034 GDIP01066339 GDIQ01086099 JAM37376.1 JAN08638.1 GDIQ01188593 GDIP01141428 JAK63132.1 JAL62286.1 GDIP01239222 JAI84179.1 GDIP01074994 JAM28721.1 GDIP01175033 GDIP01141429 GDIP01036892 JAJ48369.1 GDIP01217918 JAJ05484.1 GDIP01240376 JAI83025.1 GDIP01088483 JAM15232.1 GDIP01097840 JAM05875.1 GDIQ01087788 JAN06949.1 GDIP01067828 JAM35887.1 GDIP01074996 JAM28719.1 GDIQ01164864 JAK86861.1 GDIQ01182302 GDIQ01162357 JAK89368.1 GDIP01175036 GDIQ01163515 GDIQ01133770 JAJ48366.1 JAL17956.1 JXUM01023685 JXUM01023686 JXUM01023687 JXUM01023688 KQ560668 KXJ81360.1 GDHF01010842 JAI41472.1 GEZM01102036 JAV52042.1 GAMC01014216 JAB92339.1 KB632318 ERL92262.1 KA646463 AFP61092.1 GDHF01018034 JAI34280.1 GDHF01021822 JAI30492.1 GAMC01014215 JAB92340.1 PNF16456.1 GAKP01010684 JAC48268.1 GEZM01102035 GEZM01102034 JAV52043.1 GAKP01010687 JAC48265.1 GAKP01010683 JAC48269.1 GDHF01031101 JAI21213.1 DS235766 EEB16683.1 GBXI01014818 JAC99473.1 GDRN01077549 JAI62719.1 GL449042 EFN83313.1 GAMC01014218 JAB92337.1 ADTU01015113 KQ977574 KYN01897.1 SSX02720.1 SSX23094.1 KQ979608 KYN20374.1 GBBI01000235 JAC18477.1 GDIP01090734 JAM12981.1
Proteomes
UP000053240
UP000007151
UP000192223
UP000235965
UP000007062
UP000075883
+ More
UP000007266 UP000008820 UP000075920 UP000019118 UP000075900 UP000069272 UP000183832 UP000094527 UP000030765 UP000000673 UP000000305 UP000075882 UP000069940 UP000249989 UP000030742 UP000095301 UP000009046 UP000008237 UP000005205 UP000078542 UP000005203 UP000078492
UP000007266 UP000008820 UP000075920 UP000019118 UP000075900 UP000069272 UP000183832 UP000094527 UP000030765 UP000000673 UP000000305 UP000075882 UP000069940 UP000249989 UP000030742 UP000095301 UP000009046 UP000008237 UP000005205 UP000078542 UP000005203 UP000078492
Interpro
SUPFAM
SSF47050
SSF47050
Gene 3D
ProteinModelPortal
A0A2W1BDB1
A0A2H1W9Y7
A0A0N1IEY7
A0A212FEB5
V5GP13
A0A1W4W4N6
+ More
A0A1W4WGH3 A0A1W4WF73 A0A1W4WG72 A0A1W4W4M6 A0A2J7PJD5 A0A1W4WGG7 A0A2J7PJE1 A0A1Y1K033 Q7Q796 A0A1Y1JYF0 A0A1Y1JSP2 A0A182LTL8 D6X362 R4WQ91 Q0IEQ0 A0A1Y1JXC0 T1DCM8 A0A182WLU5 A0A2M3YY63 A0A2M4A2N4 A0A2M4A2R8 A0A2M4CY33 A0A336KE08 N6UMK8 A0A182RC20 A0A182FHN8 A0A1Q3FEV7 A0A1J1J283 U5EZV4 A0A1D2N7A2 A0A1Q3FEX1 A0A084VD99 A0A1Q3FF14 W5JD97 A0A0P5XNC5 A0A0P4XUL7 A0A0P6GFV7 A0A0P5QZ90 A0A0P5AD46 A0A0P5ZZM6 A0A0P5EKG4 A0A0P5DBB7 A0A0P5D9Z7 E9FUH3 A0A0N8A0F8 A0A0P5XYF5 A0A0P5KA17 A0A0P4XND8 A0A0P5XAD4 A0A1W4W4N2 A0A0N8A7G3 A0A0P4Z4X1 A0A1W4WF78 A0A0P4XJP7 A0A0P5WBE5 A0A0P5VMZ3 A0A0N8DVI5 A0A0P5XWA6 A0A0P5XCQ4 A0A0P5LUK6 A0A0P5NMB3 A0A0P5P4M8 A0A182LJT8 A0A182H289 A0A0K8VRG5 A0A1Y1JS46 W8BSZ4 U4UGV7 A0A1I8NEN7 T1PEY8 A0A0K8V6T8 A0A0K8UUZ2 W8AU25 A0A1I8NEP4 A0A2J7PJE0 A0A034W3D2 A0A1Y1JSQ0 A0A034W2E0 A0A034W0S3 A0A0K8U3U7 E0VTH7 A0A0A1WK92 A0A1I8NEN3 A0A1I8NEP7 A0A0P4WAZ9 E2BLU7 W8B1G0 A0A158NGA8 A0A195CN49 A0A088ABY8 A0A336LYN1 A0A195E6A0 A0A023FAJ6 A0A0P5W8X5
A0A1W4WGH3 A0A1W4WF73 A0A1W4WG72 A0A1W4W4M6 A0A2J7PJD5 A0A1W4WGG7 A0A2J7PJE1 A0A1Y1K033 Q7Q796 A0A1Y1JYF0 A0A1Y1JSP2 A0A182LTL8 D6X362 R4WQ91 Q0IEQ0 A0A1Y1JXC0 T1DCM8 A0A182WLU5 A0A2M3YY63 A0A2M4A2N4 A0A2M4A2R8 A0A2M4CY33 A0A336KE08 N6UMK8 A0A182RC20 A0A182FHN8 A0A1Q3FEV7 A0A1J1J283 U5EZV4 A0A1D2N7A2 A0A1Q3FEX1 A0A084VD99 A0A1Q3FF14 W5JD97 A0A0P5XNC5 A0A0P4XUL7 A0A0P6GFV7 A0A0P5QZ90 A0A0P5AD46 A0A0P5ZZM6 A0A0P5EKG4 A0A0P5DBB7 A0A0P5D9Z7 E9FUH3 A0A0N8A0F8 A0A0P5XYF5 A0A0P5KA17 A0A0P4XND8 A0A0P5XAD4 A0A1W4W4N2 A0A0N8A7G3 A0A0P4Z4X1 A0A1W4WF78 A0A0P4XJP7 A0A0P5WBE5 A0A0P5VMZ3 A0A0N8DVI5 A0A0P5XWA6 A0A0P5XCQ4 A0A0P5LUK6 A0A0P5NMB3 A0A0P5P4M8 A0A182LJT8 A0A182H289 A0A0K8VRG5 A0A1Y1JS46 W8BSZ4 U4UGV7 A0A1I8NEN7 T1PEY8 A0A0K8V6T8 A0A0K8UUZ2 W8AU25 A0A1I8NEP4 A0A2J7PJE0 A0A034W3D2 A0A1Y1JSQ0 A0A034W2E0 A0A034W0S3 A0A0K8U3U7 E0VTH7 A0A0A1WK92 A0A1I8NEN3 A0A1I8NEP7 A0A0P4WAZ9 E2BLU7 W8B1G0 A0A158NGA8 A0A195CN49 A0A088ABY8 A0A336LYN1 A0A195E6A0 A0A023FAJ6 A0A0P5W8X5
PDB
1V6G
E-value=1.73046e-14,
Score=193
Ontologies
GO
Topology
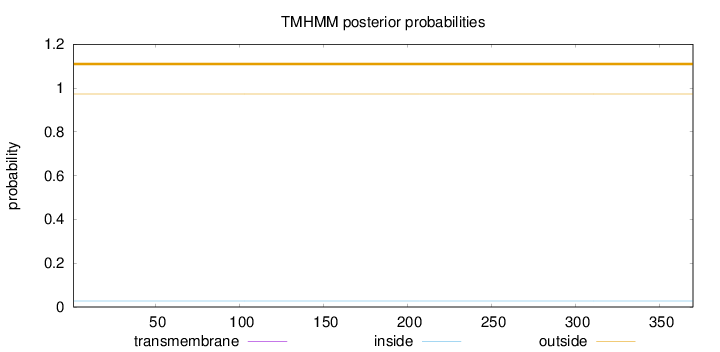
Length:
370
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00143
Exp number, first 60 AAs:
0.00128
Total prob of N-in:
0.02750
outside
1 - 370
Population Genetic Test Statistics
Pi
229.551938
Theta
166.861249
Tajima's D
1.451641
CLR
0.083847
CSRT
0.776211189440528
Interpretation
Uncertain
