Gene
KWMTBOMO15709 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002264
Annotation
PREDICTED:_actin-binding_LIM_protein_3_isoform_X3_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.504
Sequence
CDS
ATGAACAATGAGGAACCAATCGAGCTGTCACACTACCCGGCTGCTTACAAGCCGCCCCCCGGAACCAAACCGAAGATAGAGAGAGACGATTTTCCAGCACCACCTTATCCATACACAGATCCGGAACGCAGAAGGCGATGGTCGGACACGTACAAGGGCGTCCCGGACAGCGACGAGGAGGTGGACGGTAAAGTGAACGGCGCGGTCAACGGCCGGGCCGGGGTCGACCCCAAGCTGCGCAAGGAGGAGGAGGAGCTGTCCAAAATCGAGACGGGGATAGCGCAGGTGTTCCTGAAGGAAGTAAAAGAACGCGAAAAGCTTCAGCAGTGGAAGAAACAAAATTTAGATCCTAGAAACGCAAGCAGAACGCCGTCCGCGTCCCGCGAGCCGACGTACCGCCTGCGCTACTCATCGCCGGTGGGCGCGTCCCCATCCCGCAGCCTCGACCACCCGCGCGCGCTCGACGAGCCCTCGCACGCCGGCCACTCGCACGCCAGCCACTCGCACATCCCCAGCTACAACGTAATGCACGCGGCTCCGCGGCCCGGCTACGGGCTCCGGTCGAGCACGCTGCCGGCCGGCGTCGGGCGACACTGCAACGGTGACTTCACATTCAGCGGCATGGGAGATAAAACTCATAGCACCGACTTCAGCAGCGGCAAATCAGATATTTCTGCCGGCTCCATAACCGATGTGGATAGAAGTGCTGTGACAACCGAAGCGGGTATAGTGGTCCGCGGGGTGAGCACGCTGGGGTCGGGGGTGGGGGTGCGCGGCGGGGTGGGGGGCGTGCGCCGCTCCCTCCCCAACATGGCCACCTCGCATCTCCTGCACGAGCCGGCCAAGCTCTACCCCTACCACTTGCTGCTCATCACCAACTACAGGCTGCCGCCCGACGTCGACAGGCTTAACTTAGAGGTAGGTAGACGGTAG
Protein
MNNEEPIELSHYPAAYKPPPGTKPKIERDDFPAPPYPYTDPERRRRWSDTYKGVPDSDEEVDGKVNGAVNGRAGVDPKLRKEEEELSKIETGIAQVFLKEVKEREKLQQWKKQNLDPRNASRTPSASREPTYRLRYSSPVGASPSRSLDHPRALDEPSHAGHSHASHSHIPSYNVMHAAPRPGYGLRSSTLPAGVGRHCNGDFTFSGMGDKTHSTDFSSGKSDISAGSITDVDRSAVTTEAGIVVRGVSTLGSGVGVRGGVGGVRRSLPNMATSHLLHEPAKLYPYHLLLITNYRLPPDVDRLNLEVGRR
Summary
Uniprot
A0A2W1BDB1
A0A2H1W9Y7
A0A2J7PJE1
A0A1Y1JS46
A0A1Y1JYF0
A0A1Y1JSQ0
+ More
A0A1Y1JXC0 A0A067QP66 D6X362 V5GSX3 A0A2M3YY63 V5GP13 A0A2M4CY33 A0A2M4A2R8 A0A0P9BVM2 A0A2M4A2N4 A0A182SG45 B3MX65 A0A336LYN1 A0A0P8Y139 A0A336KE08 A0A1W4VMP0 A0A1W4VMW6 A0A1W4VZ77 A0A0Q9XFJ2 B4KCV9 A0A1W4W028 A0A1W4W0Q5 A0A0R1E6H6 A0A0Q9X535 Q9VH91 A0A0B4LGZ0 A0A1J1J0A3 B3NZM3 B4HJV3 A0A0R1E226 A0A0B4LGY1 B4QWH7 A0A0B4LI11 A0A1J1J283 B4JS29 B4PVC8 E1JIH2 A0A0M4F616 E1JIH3 E1JIH1 W5JD97 A0A182XCV5 A0A182HM84 Q7Q796 E0VTH7 A0A182LJT8 A0A0Q9VZM2 B4MBH9 A0A0L0CH97 A0A182IWA3 A0A182NMC6 A0A084VD99 A0A1B6I5V0 A0A0Q9X3T6 A0A182FHN8 B4GZ82 B4N9H2 A0A023EVR4 A0A3B0KQU9 A0A1B6H623 U5EZV4 A0A3B0K4L9 A0A0R3NL05 A0A3B0K6D7 B5DVQ7 A0A034W1R5 A0A3B0K1R8 A0A3B0K1Z1 A0A182VJG0 E2BLU7 A0A0K8TNM7 A0A182K422 A0A0R3NFI5 A0A034W0S3 A0A0R3NJI1 A0A0R3NFA4 A0A182WLU5 A0A1W4WF78 A0A1W4WGG7 A0A182RC20 A0A2S2Q664 A0A0A1WK92 A0A0A1X0N9 W8B1G0 A0A0P8ZL29 W8AU25 A0A182Y0R8 A0A232F716 A0A158NGA8 A0A0B4LGY0 A0A182H289 A0A3L8DXR7
A0A1Y1JXC0 A0A067QP66 D6X362 V5GSX3 A0A2M3YY63 V5GP13 A0A2M4CY33 A0A2M4A2R8 A0A0P9BVM2 A0A2M4A2N4 A0A182SG45 B3MX65 A0A336LYN1 A0A0P8Y139 A0A336KE08 A0A1W4VMP0 A0A1W4VMW6 A0A1W4VZ77 A0A0Q9XFJ2 B4KCV9 A0A1W4W028 A0A1W4W0Q5 A0A0R1E6H6 A0A0Q9X535 Q9VH91 A0A0B4LGZ0 A0A1J1J0A3 B3NZM3 B4HJV3 A0A0R1E226 A0A0B4LGY1 B4QWH7 A0A0B4LI11 A0A1J1J283 B4JS29 B4PVC8 E1JIH2 A0A0M4F616 E1JIH3 E1JIH1 W5JD97 A0A182XCV5 A0A182HM84 Q7Q796 E0VTH7 A0A182LJT8 A0A0Q9VZM2 B4MBH9 A0A0L0CH97 A0A182IWA3 A0A182NMC6 A0A084VD99 A0A1B6I5V0 A0A0Q9X3T6 A0A182FHN8 B4GZ82 B4N9H2 A0A023EVR4 A0A3B0KQU9 A0A1B6H623 U5EZV4 A0A3B0K4L9 A0A0R3NL05 A0A3B0K6D7 B5DVQ7 A0A034W1R5 A0A3B0K1R8 A0A3B0K1Z1 A0A182VJG0 E2BLU7 A0A0K8TNM7 A0A182K422 A0A0R3NFI5 A0A034W0S3 A0A0R3NJI1 A0A0R3NFA4 A0A182WLU5 A0A1W4WF78 A0A1W4WGG7 A0A182RC20 A0A2S2Q664 A0A0A1WK92 A0A0A1X0N9 W8B1G0 A0A0P8ZL29 W8AU25 A0A182Y0R8 A0A232F716 A0A158NGA8 A0A0B4LGY0 A0A182H289 A0A3L8DXR7
Pubmed
28756777
28004739
24845553
18362917
19820115
17994087
+ More
17550304 10731132 12537568 12537572 12537573 12537574 16110336 17505850 17569856 17569867 26109357 26109356 20920257 23761445 12364791 20566863 20966253 26108605 24438588 24945155 15632085 25348373 20798317 26369729 25830018 24495485 25244985 28648823 21347285 26483478 30249741
17550304 10731132 12537568 12537572 12537573 12537574 16110336 17505850 17569856 17569867 26109357 26109356 20920257 23761445 12364791 20566863 20966253 26108605 24438588 24945155 15632085 25348373 20798317 26369729 25830018 24495485 25244985 28648823 21347285 26483478 30249741
EMBL
KZ150119
PZC73219.1
ODYU01007258
SOQ49878.1
NEVH01024949
PNF16457.1
+ More
GEZM01102036 JAV52042.1 GEZM01102040 JAV52036.1 GEZM01102035 GEZM01102034 JAV52043.1 GEZM01102043 GEZM01102041 GEZM01102038 JAV52035.1 KK853179 KDR10207.1 KQ971372 EFA10334.2 GALX01005123 JAB63343.1 GGFM01000458 MBW21209.1 GALX01005124 JAB63342.1 GGFL01006039 MBW70217.1 GGFK01001710 MBW35031.1 CH902629 KPU75518.1 GGFK01001708 MBW35029.1 EDV35288.1 UFQS01000309 UFQT01000309 SSX02720.1 SSX23094.1 KPU75520.1 SSX02719.1 SSX23093.1 CH933806 KRG02424.1 EDW16981.2 CM000160 KRK03306.1 KRG02425.1 AE014297 AF434683 AY075568 AAF54427.4 AAL30427.1 AAL68375.1 AHN57256.1 CVRI01000066 CRL05885.1 CH954181 EDV49733.1 CH480815 EDW42837.1 KRK03307.1 AHN57258.1 CM000364 EDX13581.1 AHN57257.1 CRL05884.1 CH916373 EDV94569.1 EDW96701.2 ACZ94871.1 CP012526 ALC47425.1 ACZ94870.1 ACZ94872.2 ADMH02001568 ETN62046.1 APCN01004378 APCN01004379 AAAB01008960 EAA11937.4 DS235766 EEB16683.1 CH940656 KRF78297.1 EDW58450.2 JRES01000409 KNC31582.1 ATLV01011195 KE524650 KFB35943.1 GECU01025398 GECU01007142 JAS82308.1 JAT00565.1 CH964232 KRF99504.1 CH479198 EDW28100.1 EDW81648.1 GAPW01000315 JAC13283.1 OUUW01000013 SPP88246.1 GECU01037579 GECU01034952 GECU01008892 JAS70127.1 JAS72754.1 JAS98814.1 GANO01000074 JAB59797.1 SPP88243.1 CM000070 KRS99826.1 SPP88242.1 EDY67673.1 GAKP01010685 JAC48267.1 SPP88245.1 SPP88244.1 GL449042 EFN83313.1 GDAI01001877 JAI15726.1 KRS99824.1 GAKP01010683 JAC48269.1 KRS99822.1 KRS99825.1 GGMS01004001 MBY73204.1 GBXI01014818 JAC99473.1 GBXI01010084 JAD04208.1 GAMC01014218 JAB92337.1 KPU75519.1 GAMC01014215 JAB92340.1 NNAY01000793 OXU26465.1 ADTU01015113 AHN57259.1 JXUM01023685 JXUM01023686 JXUM01023687 JXUM01023688 KQ560668 KXJ81360.1 QOIP01000003 RLU25241.1
GEZM01102036 JAV52042.1 GEZM01102040 JAV52036.1 GEZM01102035 GEZM01102034 JAV52043.1 GEZM01102043 GEZM01102041 GEZM01102038 JAV52035.1 KK853179 KDR10207.1 KQ971372 EFA10334.2 GALX01005123 JAB63343.1 GGFM01000458 MBW21209.1 GALX01005124 JAB63342.1 GGFL01006039 MBW70217.1 GGFK01001710 MBW35031.1 CH902629 KPU75518.1 GGFK01001708 MBW35029.1 EDV35288.1 UFQS01000309 UFQT01000309 SSX02720.1 SSX23094.1 KPU75520.1 SSX02719.1 SSX23093.1 CH933806 KRG02424.1 EDW16981.2 CM000160 KRK03306.1 KRG02425.1 AE014297 AF434683 AY075568 AAF54427.4 AAL30427.1 AAL68375.1 AHN57256.1 CVRI01000066 CRL05885.1 CH954181 EDV49733.1 CH480815 EDW42837.1 KRK03307.1 AHN57258.1 CM000364 EDX13581.1 AHN57257.1 CRL05884.1 CH916373 EDV94569.1 EDW96701.2 ACZ94871.1 CP012526 ALC47425.1 ACZ94870.1 ACZ94872.2 ADMH02001568 ETN62046.1 APCN01004378 APCN01004379 AAAB01008960 EAA11937.4 DS235766 EEB16683.1 CH940656 KRF78297.1 EDW58450.2 JRES01000409 KNC31582.1 ATLV01011195 KE524650 KFB35943.1 GECU01025398 GECU01007142 JAS82308.1 JAT00565.1 CH964232 KRF99504.1 CH479198 EDW28100.1 EDW81648.1 GAPW01000315 JAC13283.1 OUUW01000013 SPP88246.1 GECU01037579 GECU01034952 GECU01008892 JAS70127.1 JAS72754.1 JAS98814.1 GANO01000074 JAB59797.1 SPP88243.1 CM000070 KRS99826.1 SPP88242.1 EDY67673.1 GAKP01010685 JAC48267.1 SPP88245.1 SPP88244.1 GL449042 EFN83313.1 GDAI01001877 JAI15726.1 KRS99824.1 GAKP01010683 JAC48269.1 KRS99822.1 KRS99825.1 GGMS01004001 MBY73204.1 GBXI01014818 JAC99473.1 GBXI01010084 JAD04208.1 GAMC01014218 JAB92337.1 KPU75519.1 GAMC01014215 JAB92340.1 NNAY01000793 OXU26465.1 ADTU01015113 AHN57259.1 JXUM01023685 JXUM01023686 JXUM01023687 JXUM01023688 KQ560668 KXJ81360.1 QOIP01000003 RLU25241.1
Proteomes
UP000235965
UP000027135
UP000007266
UP000007801
UP000075901
UP000192221
+ More
UP000009192 UP000002282 UP000000803 UP000183832 UP000008711 UP000001292 UP000000304 UP000001070 UP000092553 UP000000673 UP000076407 UP000075840 UP000007062 UP000009046 UP000075882 UP000008792 UP000037069 UP000075880 UP000075884 UP000030765 UP000007798 UP000069272 UP000008744 UP000268350 UP000001819 UP000075903 UP000008237 UP000075881 UP000075920 UP000192223 UP000075900 UP000076408 UP000215335 UP000005205 UP000069940 UP000249989 UP000279307
UP000009192 UP000002282 UP000000803 UP000183832 UP000008711 UP000001292 UP000000304 UP000001070 UP000092553 UP000000673 UP000076407 UP000075840 UP000007062 UP000009046 UP000075882 UP000008792 UP000037069 UP000075880 UP000075884 UP000030765 UP000007798 UP000069272 UP000008744 UP000268350 UP000001819 UP000075903 UP000008237 UP000075881 UP000075920 UP000192223 UP000075900 UP000076408 UP000215335 UP000005205 UP000069940 UP000249989 UP000279307
Interpro
SUPFAM
SSF47050
SSF47050
Gene 3D
ProteinModelPortal
A0A2W1BDB1
A0A2H1W9Y7
A0A2J7PJE1
A0A1Y1JS46
A0A1Y1JYF0
A0A1Y1JSQ0
+ More
A0A1Y1JXC0 A0A067QP66 D6X362 V5GSX3 A0A2M3YY63 V5GP13 A0A2M4CY33 A0A2M4A2R8 A0A0P9BVM2 A0A2M4A2N4 A0A182SG45 B3MX65 A0A336LYN1 A0A0P8Y139 A0A336KE08 A0A1W4VMP0 A0A1W4VMW6 A0A1W4VZ77 A0A0Q9XFJ2 B4KCV9 A0A1W4W028 A0A1W4W0Q5 A0A0R1E6H6 A0A0Q9X535 Q9VH91 A0A0B4LGZ0 A0A1J1J0A3 B3NZM3 B4HJV3 A0A0R1E226 A0A0B4LGY1 B4QWH7 A0A0B4LI11 A0A1J1J283 B4JS29 B4PVC8 E1JIH2 A0A0M4F616 E1JIH3 E1JIH1 W5JD97 A0A182XCV5 A0A182HM84 Q7Q796 E0VTH7 A0A182LJT8 A0A0Q9VZM2 B4MBH9 A0A0L0CH97 A0A182IWA3 A0A182NMC6 A0A084VD99 A0A1B6I5V0 A0A0Q9X3T6 A0A182FHN8 B4GZ82 B4N9H2 A0A023EVR4 A0A3B0KQU9 A0A1B6H623 U5EZV4 A0A3B0K4L9 A0A0R3NL05 A0A3B0K6D7 B5DVQ7 A0A034W1R5 A0A3B0K1R8 A0A3B0K1Z1 A0A182VJG0 E2BLU7 A0A0K8TNM7 A0A182K422 A0A0R3NFI5 A0A034W0S3 A0A0R3NJI1 A0A0R3NFA4 A0A182WLU5 A0A1W4WF78 A0A1W4WGG7 A0A182RC20 A0A2S2Q664 A0A0A1WK92 A0A0A1X0N9 W8B1G0 A0A0P8ZL29 W8AU25 A0A182Y0R8 A0A232F716 A0A158NGA8 A0A0B4LGY0 A0A182H289 A0A3L8DXR7
A0A1Y1JXC0 A0A067QP66 D6X362 V5GSX3 A0A2M3YY63 V5GP13 A0A2M4CY33 A0A2M4A2R8 A0A0P9BVM2 A0A2M4A2N4 A0A182SG45 B3MX65 A0A336LYN1 A0A0P8Y139 A0A336KE08 A0A1W4VMP0 A0A1W4VMW6 A0A1W4VZ77 A0A0Q9XFJ2 B4KCV9 A0A1W4W028 A0A1W4W0Q5 A0A0R1E6H6 A0A0Q9X535 Q9VH91 A0A0B4LGZ0 A0A1J1J0A3 B3NZM3 B4HJV3 A0A0R1E226 A0A0B4LGY1 B4QWH7 A0A0B4LI11 A0A1J1J283 B4JS29 B4PVC8 E1JIH2 A0A0M4F616 E1JIH3 E1JIH1 W5JD97 A0A182XCV5 A0A182HM84 Q7Q796 E0VTH7 A0A182LJT8 A0A0Q9VZM2 B4MBH9 A0A0L0CH97 A0A182IWA3 A0A182NMC6 A0A084VD99 A0A1B6I5V0 A0A0Q9X3T6 A0A182FHN8 B4GZ82 B4N9H2 A0A023EVR4 A0A3B0KQU9 A0A1B6H623 U5EZV4 A0A3B0K4L9 A0A0R3NL05 A0A3B0K6D7 B5DVQ7 A0A034W1R5 A0A3B0K1R8 A0A3B0K1Z1 A0A182VJG0 E2BLU7 A0A0K8TNM7 A0A182K422 A0A0R3NFI5 A0A034W0S3 A0A0R3NJI1 A0A0R3NFA4 A0A182WLU5 A0A1W4WF78 A0A1W4WGG7 A0A182RC20 A0A2S2Q664 A0A0A1WK92 A0A0A1X0N9 W8B1G0 A0A0P8ZL29 W8AU25 A0A182Y0R8 A0A232F716 A0A158NGA8 A0A0B4LGY0 A0A182H289 A0A3L8DXR7
Ontologies
GO
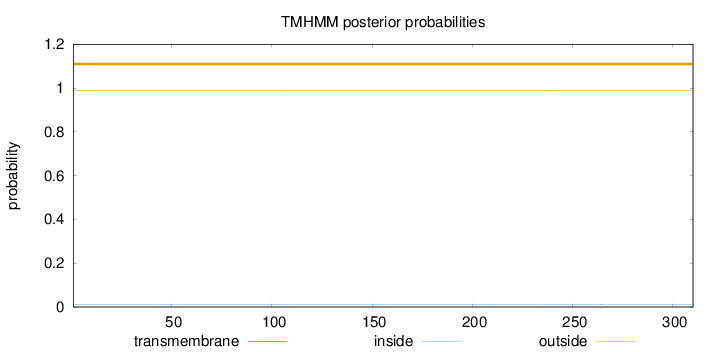
Topology
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02645
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01151
outside
1 - 310
Population Genetic Test Statistics
Pi
365.876479
Theta
207.738408
Tajima's D
2.755826
CLR
0.065257
CSRT
0.965401729913504
Interpretation
Uncertain
