Gene
KWMTBOMO15700
Pre Gene Modal
BGIBMGA002267
Annotation
PREDICTED:_cytochrome_P450_4c21-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.14
Sequence
CDS
ATGTTGATTACAGTTATAACAGACCCAGAAGACAATTTGACAATAGCCAACGGCGCACTTCAGAAACATTCTTTCTATCAGTTCGCGAGCAACTGGTTGGGGGATGGTCTCGTCACGTCAGCAGGGGAGACATGGAAGCGTCACCGCAAACTATTGAACCCAGCGTTCAGTCAGCAAATGTTAAACATCTACACAGTTGTATTCAACCAGGAATCGAAAAATCTGATTTCTGCCATCGAAATCCAAATGAAGAGCGGCCCAGTGCTTATTACCGCAGCTTTGAAAGAAATGGCATTAAAAACATTATTATCAACAGCATTCGGTATAGAAGTAGAAGAATGTGATTTCAATCAGAAGTATATGCATGCCGTTGATGAAGTAATGGCTGTGTTGACCCGTAGAATCCAGAATATTCTATTGCACAATTCGTTCGTTTACAAGCTGAGCGCTTTAAAGAAAAAGGAGGACGAGCTGATCGAAACAATAACGACGATGTCGAATAAGGTTATAAATAGTAAACGAAGAGCTTTAAAGAACAAACAGGAATCTCACGAAAATGGTTGTGCCTCTGACCTGCTCTTGAAAGGCTTGGATGACGAGGCCTTCACGGACAAAGAGATCAGGAATGAAGTCGATACCCTTATATTTACGGGGACTGATACATCGTCACAGATCATGACGGTGGTCGTGATGTTATTGGGCTCGTACCCTGAGATCCAGGATAAAGTTTACCAGGAAATTAAATCGGTGTGTGGAGATTCGGATGCCGATGTCGATAAACTTCAACATCCACGATTAGTTTACACGGAAGCTGTTTTGAAGGAAACATTAAGACTATACCCGATCACTCCGGTGGTTTTGAGAAAAACAGAAAATGAAATAAAACTGAAGAGCTACACGATACCGGCCAACAGTAACTGTATGTTGGGTATTTATGGTCTGAACCGCCACCCAGTGTGGGGCCCTGACGCGCACACGTTCCGACCAGAGCGCTGGTTGGAGCTCGGAGGTGTGCCCGACGACCCTAATGCATTCGCAGGATTCAGCGTCGGGAAACGGAATTGTATCGGTAAAACCTACGCTCTTATATCGATGAAGACCACGCTGGTACATTTATTACGACGATACAAAGTTACTGCAGATATATCTAAGATTGAATTCAAATTGGATGCGCTCATGGTGCCTTCAGACAACTGCTACGCCAAATTTGAGTCAAGAAAATGA
Protein
MLITVITDPEDNLTIANGALQKHSFYQFASNWLGDGLVTSAGETWKRHRKLLNPAFSQQMLNIYTVVFNQESKNLISAIEIQMKSGPVLITAALKEMALKTLLSTAFGIEVEECDFNQKYMHAVDEVMAVLTRRIQNILLHNSFVYKLSALKKKEDELIETITTMSNKVINSKRRALKNKQESHENGCASDLLLKGLDDEAFTDKEIRNEVDTLIFTGTDTSSQIMTVVVMLLGSYPEIQDKVYQEIKSVCGDSDADVDKLQHPRLVYTEAVLKETLRLYPITPVVLRKTENEIKLKSYTIPANSNCMLGIYGLNRHPVWGPDAHTFRPERWLELGGVPDDPNAFAGFSVGKRNCIGKTYALISMKTTLVHLLRRYKVTADISKIEFKLDALMVPSDNCYAKFESRK
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9IYD4
L0N5J5
H9IYD6
H9IYH3
L0N5K7
L0N760
+ More
A0A2A4K8W4 L0N4R8 A0A2W1BD42 A0A0N1INH0 A0A2W1BG06 A0A2A4JSD1 A0A2H1V0L7 I4DNC9 A0A2W1C018 A0A194QDS7 A0A0N0PAZ7 A0A2A4J141 A0A068EVI5 A0A2W1BKF4 A0A2A4JRL5 A0A2W1BBE6 J7FJF7 J7FJ27 A0A2W1BHI7 A0A2W1BFX5 A0A248QFF2 H9IYD5 A0A2W1BNA0 A0A2W1BHH2 A0A068F0U7 A0A194QJJ0 A0A068EU33 A0A142JYQ2 S4VAR8 A0A194QF94 A0A0N1PH37 A0A2W1BE48 A0A0K8TV47 A0A0N1IFB8 A0A248QE83 A0A2W1BG21 A0A2H1VXL2 A0A2H1VKT6 A0A2W1BLQ8 A0A2H1VM27 A0A2A4JDH2 A0A2A4IZJ1 A0A2A4IV96 A0A2A4J337 A0A2W1BXF7 A0A2W1BYK2 A0A2W1BIU6 A0A248QEI2 H9IRY4 J7FJ28 A0A2W1BTE0 H9IYD9 A0A2W1BJZ6 A0A0K8TUI0 A0A2W1BXG0 A0A2W1BIG9 A0A212EX74 A0A2W1C027 A0A194QDJ4 A0A068EWM9 A0A2W1BVD7 A0A068ETS4 A0A2W1BTD2 A0A2H1WBJ6 A0A1V0D9H3 A0A0L7L9I5 A0A2W1BTC4 A0A2W1BN87 A0A0K8TVT3 A0A2W1C2G1 A0A2H1V5V3 A0A2W1BZN2 A0A2A4JAS2 A0A212F7C8 A0A2H1VJB0 A0A2H1VHY3 A0A1V0D9H0 A0A3S2NC20 A0A2H1VI25 A0A2H1VBV8 A0A212EQP7 A0A0K8TV78 A0A2H1VFA5 A0A248QEG1 A0A346II48 J7FKN9 A0A2H1VJL3 A0A2H1V6H8 A0A1L8E485 A0A1L8E445 A0A1W6L1D9
A0A2A4K8W4 L0N4R8 A0A2W1BD42 A0A0N1INH0 A0A2W1BG06 A0A2A4JSD1 A0A2H1V0L7 I4DNC9 A0A2W1C018 A0A194QDS7 A0A0N0PAZ7 A0A2A4J141 A0A068EVI5 A0A2W1BKF4 A0A2A4JRL5 A0A2W1BBE6 J7FJF7 J7FJ27 A0A2W1BHI7 A0A2W1BFX5 A0A248QFF2 H9IYD5 A0A2W1BNA0 A0A2W1BHH2 A0A068F0U7 A0A194QJJ0 A0A068EU33 A0A142JYQ2 S4VAR8 A0A194QF94 A0A0N1PH37 A0A2W1BE48 A0A0K8TV47 A0A0N1IFB8 A0A248QE83 A0A2W1BG21 A0A2H1VXL2 A0A2H1VKT6 A0A2W1BLQ8 A0A2H1VM27 A0A2A4JDH2 A0A2A4IZJ1 A0A2A4IV96 A0A2A4J337 A0A2W1BXF7 A0A2W1BYK2 A0A2W1BIU6 A0A248QEI2 H9IRY4 J7FJ28 A0A2W1BTE0 H9IYD9 A0A2W1BJZ6 A0A0K8TUI0 A0A2W1BXG0 A0A2W1BIG9 A0A212EX74 A0A2W1C027 A0A194QDJ4 A0A068EWM9 A0A2W1BVD7 A0A068ETS4 A0A2W1BTD2 A0A2H1WBJ6 A0A1V0D9H3 A0A0L7L9I5 A0A2W1BTC4 A0A2W1BN87 A0A0K8TVT3 A0A2W1C2G1 A0A2H1V5V3 A0A2W1BZN2 A0A2A4JAS2 A0A212F7C8 A0A2H1VJB0 A0A2H1VHY3 A0A1V0D9H0 A0A3S2NC20 A0A2H1VI25 A0A2H1VBV8 A0A212EQP7 A0A0K8TV78 A0A2H1VFA5 A0A248QEG1 A0A346II48 J7FKN9 A0A2H1VJL3 A0A2H1V6H8 A0A1L8E485 A0A1L8E445 A0A1W6L1D9
Pubmed
EMBL
BABH01020614
BABH01020615
BABH01020616
BABH01020617
BABH01020618
AK289341
+ More
BAM73868.1 BABH01020606 BABH01020619 AK343194 BAM73883.1 AK343192 AK343193 BAM73881.1 NWSH01000019 PCG80725.1 AK343195 BAM73884.1 KZ150122 PZC73152.1 KQ461111 KPJ08952.1 KZ150119 PZC73211.1 NWSH01000705 PCG74676.1 ODYU01000131 SOQ34388.1 AK402852 BAM19419.1 KZ149905 PZC78316.1 KQ459144 KPJ03617.1 KPJ08951.1 NWSH01004299 PCG65244.1 KM016711 AID54863.1 PZC73210.1 PCG74677.1 KZ150169 PZC72622.1 JX310093 AFP20604.1 JX310091 AFP20602.1 PZC73154.1 PZC72625.1 KX443457 ASO98032.1 BABH01020607 BABH01020608 PZC73153.1 PZC73155.1 KM016713 AID54865.1 KPJ03616.1 KM016712 AID54864.1 KU550085 AMR93993.1 KC789755 AGO62010.1 KPJ03615.1 KPJ08953.1 PZC72621.1 GCVX01000094 JAI18136.1 KPJ08955.1 KX425014 ASN63841.1 PZC72624.1 ODYU01005067 SOQ45579.1 ODYU01003095 SOQ41416.1 PZC72623.1 ODYU01003287 SOQ41846.1 NWSH01001946 PCG69604.1 NWSH01004577 PCG64906.1 NWSH01006058 PCG63705.1 NWSH01003441 PCG66289.1 PZC78325.1 PZC78317.1 PZC73157.1 KX443458 ASO98033.1 BABH01035662 BABH01035663 JX310096 AFP20607.1 PZC78319.1 BABH01020579 BABH01020580 BABH01020581 PZC73156.1 GCVX01000093 JAI18137.1 PZC78324.1 PZC72626.1 AGBW02011819 OWR46096.1 PZC78326.1 KPJ03618.1 KM016714 AID54866.1 PZC78321.1 KM016715 AID54867.1 PZC78318.1 ODYU01007558 SOQ50433.1 KY212085 ARA91649.1 JTDY01002193 KOB71906.1 PZC78322.1 KZ150062 PZC74190.1 GCVX01000055 JAI18175.1 PZC78323.1 ODYU01000849 SOQ36230.1 PZC78320.1 NWSH01002233 PCG68866.1 AGBW02009886 OWR49643.1 ODYU01002870 SOQ40886.1 ODYU01002657 SOQ40445.1 KY212086 ARA91650.1 RSAL01000110 RVE47143.1 SOQ40446.1 ODYU01001698 SOQ38276.1 AGBW02013250 OWR43823.1 GCVX01000064 JAI18166.1 ODYU01001992 SOQ38944.1 KX425017 ASN63844.1 MH138262 AXP17203.1 JX310097 AFP20608.1 SOQ40444.1 ODYU01000929 SOQ36438.1 GFDF01000640 JAV13444.1 GFDF01000581 JAV13503.1 KX609528 ARN17935.1
BAM73868.1 BABH01020606 BABH01020619 AK343194 BAM73883.1 AK343192 AK343193 BAM73881.1 NWSH01000019 PCG80725.1 AK343195 BAM73884.1 KZ150122 PZC73152.1 KQ461111 KPJ08952.1 KZ150119 PZC73211.1 NWSH01000705 PCG74676.1 ODYU01000131 SOQ34388.1 AK402852 BAM19419.1 KZ149905 PZC78316.1 KQ459144 KPJ03617.1 KPJ08951.1 NWSH01004299 PCG65244.1 KM016711 AID54863.1 PZC73210.1 PCG74677.1 KZ150169 PZC72622.1 JX310093 AFP20604.1 JX310091 AFP20602.1 PZC73154.1 PZC72625.1 KX443457 ASO98032.1 BABH01020607 BABH01020608 PZC73153.1 PZC73155.1 KM016713 AID54865.1 KPJ03616.1 KM016712 AID54864.1 KU550085 AMR93993.1 KC789755 AGO62010.1 KPJ03615.1 KPJ08953.1 PZC72621.1 GCVX01000094 JAI18136.1 KPJ08955.1 KX425014 ASN63841.1 PZC72624.1 ODYU01005067 SOQ45579.1 ODYU01003095 SOQ41416.1 PZC72623.1 ODYU01003287 SOQ41846.1 NWSH01001946 PCG69604.1 NWSH01004577 PCG64906.1 NWSH01006058 PCG63705.1 NWSH01003441 PCG66289.1 PZC78325.1 PZC78317.1 PZC73157.1 KX443458 ASO98033.1 BABH01035662 BABH01035663 JX310096 AFP20607.1 PZC78319.1 BABH01020579 BABH01020580 BABH01020581 PZC73156.1 GCVX01000093 JAI18137.1 PZC78324.1 PZC72626.1 AGBW02011819 OWR46096.1 PZC78326.1 KPJ03618.1 KM016714 AID54866.1 PZC78321.1 KM016715 AID54867.1 PZC78318.1 ODYU01007558 SOQ50433.1 KY212085 ARA91649.1 JTDY01002193 KOB71906.1 PZC78322.1 KZ150062 PZC74190.1 GCVX01000055 JAI18175.1 PZC78323.1 ODYU01000849 SOQ36230.1 PZC78320.1 NWSH01002233 PCG68866.1 AGBW02009886 OWR49643.1 ODYU01002870 SOQ40886.1 ODYU01002657 SOQ40445.1 KY212086 ARA91650.1 RSAL01000110 RVE47143.1 SOQ40446.1 ODYU01001698 SOQ38276.1 AGBW02013250 OWR43823.1 GCVX01000064 JAI18166.1 ODYU01001992 SOQ38944.1 KX425017 ASN63844.1 MH138262 AXP17203.1 JX310097 AFP20608.1 SOQ40444.1 ODYU01000929 SOQ36438.1 GFDF01000640 JAV13444.1 GFDF01000581 JAV13503.1 KX609528 ARN17935.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
H9IYD4
L0N5J5
H9IYD6
H9IYH3
L0N5K7
L0N760
+ More
A0A2A4K8W4 L0N4R8 A0A2W1BD42 A0A0N1INH0 A0A2W1BG06 A0A2A4JSD1 A0A2H1V0L7 I4DNC9 A0A2W1C018 A0A194QDS7 A0A0N0PAZ7 A0A2A4J141 A0A068EVI5 A0A2W1BKF4 A0A2A4JRL5 A0A2W1BBE6 J7FJF7 J7FJ27 A0A2W1BHI7 A0A2W1BFX5 A0A248QFF2 H9IYD5 A0A2W1BNA0 A0A2W1BHH2 A0A068F0U7 A0A194QJJ0 A0A068EU33 A0A142JYQ2 S4VAR8 A0A194QF94 A0A0N1PH37 A0A2W1BE48 A0A0K8TV47 A0A0N1IFB8 A0A248QE83 A0A2W1BG21 A0A2H1VXL2 A0A2H1VKT6 A0A2W1BLQ8 A0A2H1VM27 A0A2A4JDH2 A0A2A4IZJ1 A0A2A4IV96 A0A2A4J337 A0A2W1BXF7 A0A2W1BYK2 A0A2W1BIU6 A0A248QEI2 H9IRY4 J7FJ28 A0A2W1BTE0 H9IYD9 A0A2W1BJZ6 A0A0K8TUI0 A0A2W1BXG0 A0A2W1BIG9 A0A212EX74 A0A2W1C027 A0A194QDJ4 A0A068EWM9 A0A2W1BVD7 A0A068ETS4 A0A2W1BTD2 A0A2H1WBJ6 A0A1V0D9H3 A0A0L7L9I5 A0A2W1BTC4 A0A2W1BN87 A0A0K8TVT3 A0A2W1C2G1 A0A2H1V5V3 A0A2W1BZN2 A0A2A4JAS2 A0A212F7C8 A0A2H1VJB0 A0A2H1VHY3 A0A1V0D9H0 A0A3S2NC20 A0A2H1VI25 A0A2H1VBV8 A0A212EQP7 A0A0K8TV78 A0A2H1VFA5 A0A248QEG1 A0A346II48 J7FKN9 A0A2H1VJL3 A0A2H1V6H8 A0A1L8E485 A0A1L8E445 A0A1W6L1D9
A0A2A4K8W4 L0N4R8 A0A2W1BD42 A0A0N1INH0 A0A2W1BG06 A0A2A4JSD1 A0A2H1V0L7 I4DNC9 A0A2W1C018 A0A194QDS7 A0A0N0PAZ7 A0A2A4J141 A0A068EVI5 A0A2W1BKF4 A0A2A4JRL5 A0A2W1BBE6 J7FJF7 J7FJ27 A0A2W1BHI7 A0A2W1BFX5 A0A248QFF2 H9IYD5 A0A2W1BNA0 A0A2W1BHH2 A0A068F0U7 A0A194QJJ0 A0A068EU33 A0A142JYQ2 S4VAR8 A0A194QF94 A0A0N1PH37 A0A2W1BE48 A0A0K8TV47 A0A0N1IFB8 A0A248QE83 A0A2W1BG21 A0A2H1VXL2 A0A2H1VKT6 A0A2W1BLQ8 A0A2H1VM27 A0A2A4JDH2 A0A2A4IZJ1 A0A2A4IV96 A0A2A4J337 A0A2W1BXF7 A0A2W1BYK2 A0A2W1BIU6 A0A248QEI2 H9IRY4 J7FJ28 A0A2W1BTE0 H9IYD9 A0A2W1BJZ6 A0A0K8TUI0 A0A2W1BXG0 A0A2W1BIG9 A0A212EX74 A0A2W1C027 A0A194QDJ4 A0A068EWM9 A0A2W1BVD7 A0A068ETS4 A0A2W1BTD2 A0A2H1WBJ6 A0A1V0D9H3 A0A0L7L9I5 A0A2W1BTC4 A0A2W1BN87 A0A0K8TVT3 A0A2W1C2G1 A0A2H1V5V3 A0A2W1BZN2 A0A2A4JAS2 A0A212F7C8 A0A2H1VJB0 A0A2H1VHY3 A0A1V0D9H0 A0A3S2NC20 A0A2H1VI25 A0A2H1VBV8 A0A212EQP7 A0A0K8TV78 A0A2H1VFA5 A0A248QEG1 A0A346II48 J7FKN9 A0A2H1VJL3 A0A2H1V6H8 A0A1L8E485 A0A1L8E445 A0A1W6L1D9
PDB
6C93
E-value=1.76962e-43,
Score=443
Ontologies
GO
Topology
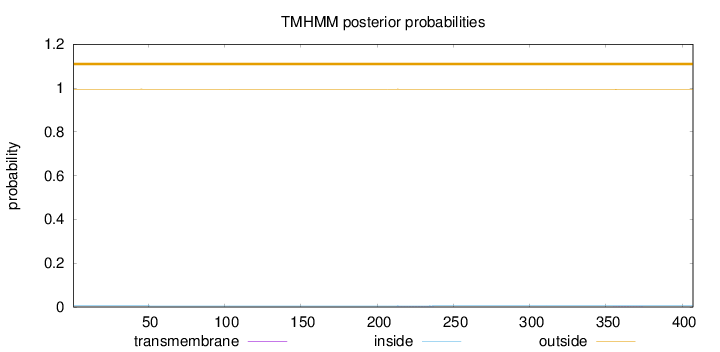
Length:
407
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03175
Exp number, first 60 AAs:
0.00231
Total prob of N-in:
0.00520
outside
1 - 407
Population Genetic Test Statistics
Pi
195.4615
Theta
164.432351
Tajima's D
0.161512
CLR
2.481707
CSRT
0.412329383530823
Interpretation
Uncertain
