Gene
KWMTBOMO15692
Pre Gene Modal
BGIBMGA002273
Annotation
PREDICTED:_wolframin_isoform_X4_[Bombyx_mori]
Full name
Wolframin
Location in the cell
PlasmaMembrane Reliability : 4.671
Sequence
CDS
ATGCCCGGGGGACGCAAACGGTGGAATTTACATGATGGACCTCAAGGGTCACTACGTCGACTTAGGAATCAGTTAGCTGAGGATGGTTGTGCTGAATCTCAAGTGGTACTGGCTAAACAGCTCTTGGAAGAAAAATGTGAGCTTGAGGCTGACAAGATATCAAATTTCAAACAGGCTTTAGAATGGCTCATATGCGCAACGGAACAAGCACACCCGGAAGCAAGGAGGCTTTTAAGAAGGTGTATCCGTAGCGGCGTCATAGATGAGGACAGCGCAGCGATAGTACGTGCCAAGTCTTGTCTGGCGGCTAGCAGACAGGAGACAGTCGCTAGGAAAGCGGCTAGAGATCTATTTGCCAGTTTGTCGAATGGTGAACAATACATAACAACGGTTCAACTCGAACGTCGGATAAGAGAGATTTGCAATACGACGTTACGGAAGACAGCGGCCGAAGACAGCGATTCTCCTGACGACGATAACTCAGAAGCGGCAAATGCGCTAGACGATGCCGCACAAGCATTAGAACCATCTGACCTACAAGCTGGCGATCACATAAACAGTAATGGAACGCTGCGTACCTCACATGACCTCGAAGAAGATGTACCTGACAGATGTTCGAATGAAGAGATACGTAACTTGACAGTTGACAATCTAGTATCGGCAGCGGTCGATTATTGTCAGGGCGAACTGCCTCTAGTCAGCTATGAATTGACCTTAACGGACCCTAGCGTTAAGTCTCTCGATCACATTCCATTGCTTCAACAAGCCTTTCTACATCCAATAGTTTTCTTACAAGTTTTATATCTCAAACTACTCTATTACTTTGGTTCGTTTTCGTTTAGATTATCCAACATAGAGCTGTCTTTAGTTCTAATAGCGTATCTATTCGTTAGTGCCGATAGTTTGTATCATTTAGTTCCGCTAGTCCTTTACTACGTCAGTTTCGTCGCCATGGTGATTTGCACGTTCAAAATGTTGCATGCGAAACGACAGTTTATCGATTTTCGTAAGTGGTCCGGTCTCTTCTTGAGGTACAGCGACGGTAATTTGCAACCAGACGAGTCTGAGAATTTGTTTGTACGCAACAATCTGGGTCCGTTCATTCAGTTCTTTGTGGCGTTGTTTGTGAATTTATTTTTATATCCCTTTATAGCAACACAATGGGTTCCGTTTTCTGAGCTCTGCGTGTTGTCCTTCTTTCTGATGTTCCTTACACTGTGTTCATTTGGAGCTAACGGGAACCCTTATCCGGACGTTTTAGCTCTAATTTCATTCGCCATTAACGTGCTGGCCAAATACCCCTACGAGAAGGATACAGTTGTACATCAGGGGTGGAGATTTTTAGATTTTCACATATCGAATTATCCGTCTTATGTCCTCGGGAATAGCATAGAGTTTTGTTTGAACGGTCGAATGTTCTTTTCGTTGTTGATACCGGTCATTTTGGTGTTGATGGCGCGCAGAAGCGATTGGCAGGGTTTTTTTAAACAAACGCTACCACATTGCGTGACTTTAAGCTGGTTGCAGATGTTCATAATGTCTTCACATGGCTGTACCACGTACGGATTGATTAGGGGGACATTGGCGTTAGTGTGTTCATTCCTATTTCTTCCATTAATCGGAGTGATGACCGTGACCTTGCCATTATTCGCGTTTCTGCAGTACATTACGTTGCCAAGGATTATTTATACGGCGACCATGGTGTTGAGCTTTGGCCTAAGTTTGGGCTTTACTTGTTTGCTGGCAAAGTCTGAAGCAACGAAGCGATTTGTCACTCCGTTTCAGCTCGTATTCGGTACATTAGCCATTATATACTTTGGGAACCAGTTCGTTTTGAACATACGCGATGACGGTCTCCCGTCAGGCCTTATCGAACTATTGGCCGATGAGAAGCCGAGCATAAAGAACCTTCTAAAAAACGAATTCCTTAGCGAATACGAAGACAACAATTACCACATAAATTGGGACGATTATTACACTCACTGTAACAATCCATCTTGGACTGATTATAATATAGCCGCAACTCAAATCAAGTGTTCGGTACTAGAGGGCGCGAACGTCAACTGGGAAGGTTATGTCAAAGATGTCAGTTTATTAAACGTCAAAAATAAATGGAGGGACTTTGCGTATTGGTTGCCGGGTTTCTTGCAAGAGTACTTTAAGTGCTATTACGGCGAGGAGTATTCGAATGTGTGTCGAGGCTCGGACGGTGTTCAGTCTGTGGCGGAATGCGAATTCGTTAGCAGTGTTGCCAGGCAAAGCGGAAAGAGTTGCCATCTTGATAACTTGAATGAATACACGTATGAAGTGAAATTAATGATGGAAGCCAGTGGCGGTATTTTGAAGCGCCACGCGGAAATCGTCATACACTTTGGTCATTATTTTACGAATTTTACACGTTTACTGCGATCGGACGACAAGATACGTTTTAAAGGGACGCTTTCGAACGAAAACCGACCTTATAACATTGGCGCTAAGGACTTGACTATAAGGGGCTACGAAATTAACTGTTTGGAATGCAAAGAGGCGCGCGGCTCTGTGACTCTGAAGACGTCGTCCGCATCGAAATTCTCTGAGCTGTTTAAAACAATTGTGAATGATTGCGTTGTATCGACAAAGTATATTTTGAATTTCATGTTGAATCCTATAATTGTTTTCAAATAA
Protein
MPGGRKRWNLHDGPQGSLRRLRNQLAEDGCAESQVVLAKQLLEEKCELEADKISNFKQALEWLICATEQAHPEARRLLRRCIRSGVIDEDSAAIVRAKSCLAASRQETVARKAARDLFASLSNGEQYITTVQLERRIREICNTTLRKTAAEDSDSPDDDNSEAANALDDAAQALEPSDLQAGDHINSNGTLRTSHDLEEDVPDRCSNEEIRNLTVDNLVSAAVDYCQGELPLVSYELTLTDPSVKSLDHIPLLQQAFLHPIVFLQVLYLKLLYYFGSFSFRLSNIELSLVLIAYLFVSADSLYHLVPLVLYYVSFVAMVICTFKMLHAKRQFIDFRKWSGLFLRYSDGNLQPDESENLFVRNNLGPFIQFFVALFVNLFLYPFIATQWVPFSELCVLSFFLMFLTLCSFGANGNPYPDVLALISFAINVLAKYPYEKDTVVHQGWRFLDFHISNYPSYVLGNSIEFCLNGRMFFSLLIPVILVLMARRSDWQGFFKQTLPHCVTLSWLQMFIMSSHGCTTYGLIRGTLALVCSFLFLPLIGVMTVTLPLFAFLQYITLPRIIYTATMVLSFGLSLGFTCLLAKSEATKRFVTPFQLVFGTLAIIYFGNQFVLNIRDDGLPSGLIELLADEKPSIKNLLKNEFLSEYEDNNYHINWDDYYTHCNNPSWTDYNIAATQIKCSVLEGANVNWEGYVKDVSLLNVKNKWRDFAYWLPGFLQEYFKCYYGEEYSNVCRGSDGVQSVAECEFVSSVARQSGKSCHLDNLNEYTYEVKLMMEASGGILKRHAEIVIHFGHYFTNFTRLLRSDDKIRFKGTLSNENRPYNIGAKDLTIRGYEINCLECKEARGSVTLKTSSASKFSELFKTIVNDCVVSTKYILNFMLNPIIVFK
Summary
Description
Participates in the regulation of cellular Ca(2+) homeostasis, at least partly, by modulating the filling state of the endoplasmic reticulum Ca(2+) store (By similarity). In neurons and glial cells, has a role in maintaining neuronal function and integrity during aging (PubMed:29357349).
Keywords
Alternative splicing
Complete proteome
Endoplasmic reticulum
Glycoprotein
Membrane
Mitochondrion
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Wolframin
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9IYE0
A0A2W1BD50
A0A2H1VKQ0
A0A194QF89
A0A0N1I708
A0A1E1WGH4
+ More
A0A212F968 A0A0L7LDZ0 D6X0P3 A0A0L7QL14 A0A088AFE1 A0A0T6AW23 A0A1Y1LW29 E2A2Z7 A0A158NXQ4 F4X4B3 A0A2J7PI79 A0A0N0BD29 A0A1Y1LXD6 A0A067R4P7 A0A195BSD6 A0A2A3ED75 A0A3L8DZL1 A0A195E4H1 A0A151WR02 A0A195FHM7 B0X167 A0A151I6F5 A0A154PSY5 A0A026W9B5 E9ICB1 A0A1Q3F2T7 Q171Y3 A0A182GCP4 A0A182G1I2 A0A0N8ES65 A0A1S4FGN5 A0A336M988 E2BH31 A0A1Y1LZD7 A0A0C9QKN0 A0A0L0C7H6 A0A1I8Q0D4 K7J8R9 A0A1I8MVQ3 T1P809 A0A182Q5F5 A0A182JCT5 A0A1A9ZLD1 A0A1A9YIX8 A0A182RI24 Q7QA69 A0A182UNT3 A0A1B0C0Y0 A0A182HVN9 A0A182K4W4 A0A182KZS0 A0A232EZB0 A0A2M4BDH5 A0A182XF68 A0A2M4BCX6 A0A182UB07 A0A1A9VZX8 A0A182FAF5 A0A182Y907 U5EYL1 A0A182T659 A0A182NCV0 A0A2M4AMS2 A0A336MAJ0 A0A182W385 A0A182M7N0 W5JSX2 A0A0A1X9P0 A0A0K8U828 A0A034WSW1 A0A1A9VQX9 A0A182PLH1 Q8IMZ9-2 A0A0R1E4B4 B3P8L4 W8BU78 U4TXE6 Q8IMZ9 B4PMD5 N6TMB5 B4R0P1 A0A084VDJ9 A0A1W4VL02 B4HER8 B3M366 B4NBR7 A0A0M5J5I5 B4JRZ8 B4M642 A0A1B0GMR6 N6UKG0 A0A3B0KUZ2
A0A212F968 A0A0L7LDZ0 D6X0P3 A0A0L7QL14 A0A088AFE1 A0A0T6AW23 A0A1Y1LW29 E2A2Z7 A0A158NXQ4 F4X4B3 A0A2J7PI79 A0A0N0BD29 A0A1Y1LXD6 A0A067R4P7 A0A195BSD6 A0A2A3ED75 A0A3L8DZL1 A0A195E4H1 A0A151WR02 A0A195FHM7 B0X167 A0A151I6F5 A0A154PSY5 A0A026W9B5 E9ICB1 A0A1Q3F2T7 Q171Y3 A0A182GCP4 A0A182G1I2 A0A0N8ES65 A0A1S4FGN5 A0A336M988 E2BH31 A0A1Y1LZD7 A0A0C9QKN0 A0A0L0C7H6 A0A1I8Q0D4 K7J8R9 A0A1I8MVQ3 T1P809 A0A182Q5F5 A0A182JCT5 A0A1A9ZLD1 A0A1A9YIX8 A0A182RI24 Q7QA69 A0A182UNT3 A0A1B0C0Y0 A0A182HVN9 A0A182K4W4 A0A182KZS0 A0A232EZB0 A0A2M4BDH5 A0A182XF68 A0A2M4BCX6 A0A182UB07 A0A1A9VZX8 A0A182FAF5 A0A182Y907 U5EYL1 A0A182T659 A0A182NCV0 A0A2M4AMS2 A0A336MAJ0 A0A182W385 A0A182M7N0 W5JSX2 A0A0A1X9P0 A0A0K8U828 A0A034WSW1 A0A1A9VQX9 A0A182PLH1 Q8IMZ9-2 A0A0R1E4B4 B3P8L4 W8BU78 U4TXE6 Q8IMZ9 B4PMD5 N6TMB5 B4R0P1 A0A084VDJ9 A0A1W4VL02 B4HER8 B3M366 B4NBR7 A0A0M5J5I5 B4JRZ8 B4M642 A0A1B0GMR6 N6UKG0 A0A3B0KUZ2
Pubmed
19121390
28756777
26354079
22118469
26227816
18362917
+ More
19820115 28004739 20798317 21347285 21719571 24845553 30249741 24508170 21282665 17510324 26483478 26999592 26108605 20075255 25315136 12364791 14747013 17210077 20966253 28648823 25244985 20920257 23761445 25830018 25348373 10731132 12537572 12537569 29357349 17994087 17550304 24495485 23537049 24438588
19820115 28004739 20798317 21347285 21719571 24845553 30249741 24508170 21282665 17510324 26483478 26999592 26108605 20075255 25315136 12364791 14747013 17210077 20966253 28648823 25244985 20920257 23761445 25830018 25348373 10731132 12537572 12537569 29357349 17994087 17550304 24495485 23537049 24438588
EMBL
BABH01020573
BABH01020574
BABH01020575
BABH01020576
KZ150122
PZC73169.1
+ More
ODYU01003100 SOQ41429.1 KQ459144 KPJ03610.1 KQ461111 KPJ08944.1 GDQN01004996 JAT86058.1 AGBW02009638 OWR50271.1 JTDY01001491 KOB73743.1 KQ971371 EFA10125.2 KQ414940 KOC59221.1 LJIG01022687 KRT79249.1 GEZM01045539 JAV77759.1 GL436239 EFN72205.1 ADTU01000477 GL888633 EGI58709.1 NEVH01025129 PNF16038.1 KQ435879 KOX69868.1 GEZM01045537 JAV77761.1 KK852895 KDR14192.1 KQ976423 KYM88917.1 KZ288287 PBC29434.1 QOIP01000002 RLU25786.1 KQ979685 KYN19802.1 KQ982815 KYQ50238.1 KQ981606 KYN39509.1 DS232254 EDS38516.1 KQ978476 KYM93679.1 KQ435180 KZC15012.1 KK107323 EZA52583.1 GL762231 EFZ21806.1 GFDL01013237 JAV21808.1 CH477443 EAT40803.1 JXUM01054530 KQ561824 KXJ77421.1 JXUM01023445 JXUM01023446 KQ560661 KXJ81387.1 GDUN01000330 JAN95589.1 UFQT01000564 SSX25353.1 GL448268 EFN84952.1 GEZM01045538 JAV77760.1 GBYB01002846 GBYB01004109 GBYB01004254 JAG72613.1 JAG73876.1 JAG74021.1 JRES01000809 KNC28230.1 KA644771 AFP59400.1 AXCN02001206 AAAB01008898 EAA09198.5 JXJN01023809 APCN01005808 NNAY01001520 OXU23705.1 GGFJ01001717 MBW50858.1 GGFJ01001716 MBW50857.1 GANO01001938 JAB57933.1 GGFK01008775 MBW42096.1 UFQT01000793 SSX27276.1 AXCM01000104 ADMH02000180 ETN67462.1 GBXI01008183 GBXI01006273 JAD06109.1 JAD08019.1 GDHF01029794 JAI22520.1 GAKP01002084 JAC56868.1 AE014297 AY119144 AAM51004.1 CM000160 KRK04091.1 CH954182 EDV54109.1 GAMC01009699 JAB96856.1 KB631805 ERL86279.1 EDW98040.1 APGK01029501 KB740735 ENN79168.1 CM000364 EDX13964.1 ATLV01011556 KE524695 KFB36043.1 CH480815 EDW43229.1 CH902617 EDV42466.1 CH964232 EDW81231.1 CP012526 ALC47781.1 CH916373 EDV94538.1 CH940652 EDW59118.1 AJVK01026430 AJVK01026431 AJVK01026432 ENN79167.1 OUUW01000013 SPP87768.1
ODYU01003100 SOQ41429.1 KQ459144 KPJ03610.1 KQ461111 KPJ08944.1 GDQN01004996 JAT86058.1 AGBW02009638 OWR50271.1 JTDY01001491 KOB73743.1 KQ971371 EFA10125.2 KQ414940 KOC59221.1 LJIG01022687 KRT79249.1 GEZM01045539 JAV77759.1 GL436239 EFN72205.1 ADTU01000477 GL888633 EGI58709.1 NEVH01025129 PNF16038.1 KQ435879 KOX69868.1 GEZM01045537 JAV77761.1 KK852895 KDR14192.1 KQ976423 KYM88917.1 KZ288287 PBC29434.1 QOIP01000002 RLU25786.1 KQ979685 KYN19802.1 KQ982815 KYQ50238.1 KQ981606 KYN39509.1 DS232254 EDS38516.1 KQ978476 KYM93679.1 KQ435180 KZC15012.1 KK107323 EZA52583.1 GL762231 EFZ21806.1 GFDL01013237 JAV21808.1 CH477443 EAT40803.1 JXUM01054530 KQ561824 KXJ77421.1 JXUM01023445 JXUM01023446 KQ560661 KXJ81387.1 GDUN01000330 JAN95589.1 UFQT01000564 SSX25353.1 GL448268 EFN84952.1 GEZM01045538 JAV77760.1 GBYB01002846 GBYB01004109 GBYB01004254 JAG72613.1 JAG73876.1 JAG74021.1 JRES01000809 KNC28230.1 KA644771 AFP59400.1 AXCN02001206 AAAB01008898 EAA09198.5 JXJN01023809 APCN01005808 NNAY01001520 OXU23705.1 GGFJ01001717 MBW50858.1 GGFJ01001716 MBW50857.1 GANO01001938 JAB57933.1 GGFK01008775 MBW42096.1 UFQT01000793 SSX27276.1 AXCM01000104 ADMH02000180 ETN67462.1 GBXI01008183 GBXI01006273 JAD06109.1 JAD08019.1 GDHF01029794 JAI22520.1 GAKP01002084 JAC56868.1 AE014297 AY119144 AAM51004.1 CM000160 KRK04091.1 CH954182 EDV54109.1 GAMC01009699 JAB96856.1 KB631805 ERL86279.1 EDW98040.1 APGK01029501 KB740735 ENN79168.1 CM000364 EDX13964.1 ATLV01011556 KE524695 KFB36043.1 CH480815 EDW43229.1 CH902617 EDV42466.1 CH964232 EDW81231.1 CP012526 ALC47781.1 CH916373 EDV94538.1 CH940652 EDW59118.1 AJVK01026430 AJVK01026431 AJVK01026432 ENN79167.1 OUUW01000013 SPP87768.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000007266
+ More
UP000053825 UP000005203 UP000000311 UP000005205 UP000007755 UP000235965 UP000053105 UP000027135 UP000078540 UP000242457 UP000279307 UP000078492 UP000075809 UP000078541 UP000002320 UP000078542 UP000076502 UP000053097 UP000008820 UP000069940 UP000249989 UP000008237 UP000037069 UP000095300 UP000002358 UP000095301 UP000075886 UP000075880 UP000092445 UP000092443 UP000075900 UP000007062 UP000075903 UP000092460 UP000075840 UP000075881 UP000075882 UP000215335 UP000076407 UP000075902 UP000091820 UP000069272 UP000076408 UP000075901 UP000075884 UP000075920 UP000075883 UP000000673 UP000078200 UP000075885 UP000000803 UP000002282 UP000008711 UP000030742 UP000019118 UP000000304 UP000030765 UP000192221 UP000001292 UP000007801 UP000007798 UP000092553 UP000001070 UP000008792 UP000092462 UP000268350
UP000053825 UP000005203 UP000000311 UP000005205 UP000007755 UP000235965 UP000053105 UP000027135 UP000078540 UP000242457 UP000279307 UP000078492 UP000075809 UP000078541 UP000002320 UP000078542 UP000076502 UP000053097 UP000008820 UP000069940 UP000249989 UP000008237 UP000037069 UP000095300 UP000002358 UP000095301 UP000075886 UP000075880 UP000092445 UP000092443 UP000075900 UP000007062 UP000075903 UP000092460 UP000075840 UP000075881 UP000075882 UP000215335 UP000076407 UP000075902 UP000091820 UP000069272 UP000076408 UP000075901 UP000075884 UP000075920 UP000075883 UP000000673 UP000078200 UP000075885 UP000000803 UP000002282 UP000008711 UP000030742 UP000019118 UP000000304 UP000030765 UP000192221 UP000001292 UP000007801 UP000007798 UP000092553 UP000001070 UP000008792 UP000092462 UP000268350
Pfam
PF05510 Sarcoglycan_2
Interpro
SUPFAM
SSF49313
SSF49313
Gene 3D
ProteinModelPortal
H9IYE0
A0A2W1BD50
A0A2H1VKQ0
A0A194QF89
A0A0N1I708
A0A1E1WGH4
+ More
A0A212F968 A0A0L7LDZ0 D6X0P3 A0A0L7QL14 A0A088AFE1 A0A0T6AW23 A0A1Y1LW29 E2A2Z7 A0A158NXQ4 F4X4B3 A0A2J7PI79 A0A0N0BD29 A0A1Y1LXD6 A0A067R4P7 A0A195BSD6 A0A2A3ED75 A0A3L8DZL1 A0A195E4H1 A0A151WR02 A0A195FHM7 B0X167 A0A151I6F5 A0A154PSY5 A0A026W9B5 E9ICB1 A0A1Q3F2T7 Q171Y3 A0A182GCP4 A0A182G1I2 A0A0N8ES65 A0A1S4FGN5 A0A336M988 E2BH31 A0A1Y1LZD7 A0A0C9QKN0 A0A0L0C7H6 A0A1I8Q0D4 K7J8R9 A0A1I8MVQ3 T1P809 A0A182Q5F5 A0A182JCT5 A0A1A9ZLD1 A0A1A9YIX8 A0A182RI24 Q7QA69 A0A182UNT3 A0A1B0C0Y0 A0A182HVN9 A0A182K4W4 A0A182KZS0 A0A232EZB0 A0A2M4BDH5 A0A182XF68 A0A2M4BCX6 A0A182UB07 A0A1A9VZX8 A0A182FAF5 A0A182Y907 U5EYL1 A0A182T659 A0A182NCV0 A0A2M4AMS2 A0A336MAJ0 A0A182W385 A0A182M7N0 W5JSX2 A0A0A1X9P0 A0A0K8U828 A0A034WSW1 A0A1A9VQX9 A0A182PLH1 Q8IMZ9-2 A0A0R1E4B4 B3P8L4 W8BU78 U4TXE6 Q8IMZ9 B4PMD5 N6TMB5 B4R0P1 A0A084VDJ9 A0A1W4VL02 B4HER8 B3M366 B4NBR7 A0A0M5J5I5 B4JRZ8 B4M642 A0A1B0GMR6 N6UKG0 A0A3B0KUZ2
A0A212F968 A0A0L7LDZ0 D6X0P3 A0A0L7QL14 A0A088AFE1 A0A0T6AW23 A0A1Y1LW29 E2A2Z7 A0A158NXQ4 F4X4B3 A0A2J7PI79 A0A0N0BD29 A0A1Y1LXD6 A0A067R4P7 A0A195BSD6 A0A2A3ED75 A0A3L8DZL1 A0A195E4H1 A0A151WR02 A0A195FHM7 B0X167 A0A151I6F5 A0A154PSY5 A0A026W9B5 E9ICB1 A0A1Q3F2T7 Q171Y3 A0A182GCP4 A0A182G1I2 A0A0N8ES65 A0A1S4FGN5 A0A336M988 E2BH31 A0A1Y1LZD7 A0A0C9QKN0 A0A0L0C7H6 A0A1I8Q0D4 K7J8R9 A0A1I8MVQ3 T1P809 A0A182Q5F5 A0A182JCT5 A0A1A9ZLD1 A0A1A9YIX8 A0A182RI24 Q7QA69 A0A182UNT3 A0A1B0C0Y0 A0A182HVN9 A0A182K4W4 A0A182KZS0 A0A232EZB0 A0A2M4BDH5 A0A182XF68 A0A2M4BCX6 A0A182UB07 A0A1A9VZX8 A0A182FAF5 A0A182Y907 U5EYL1 A0A182T659 A0A182NCV0 A0A2M4AMS2 A0A336MAJ0 A0A182W385 A0A182M7N0 W5JSX2 A0A0A1X9P0 A0A0K8U828 A0A034WSW1 A0A1A9VQX9 A0A182PLH1 Q8IMZ9-2 A0A0R1E4B4 B3P8L4 W8BU78 U4TXE6 Q8IMZ9 B4PMD5 N6TMB5 B4R0P1 A0A084VDJ9 A0A1W4VL02 B4HER8 B3M366 B4NBR7 A0A0M5J5I5 B4JRZ8 B4M642 A0A1B0GMR6 N6UKG0 A0A3B0KUZ2
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
Endoplasmic reticulum
Mitochondrion
Endoplasmic reticulum
Mitochondrion
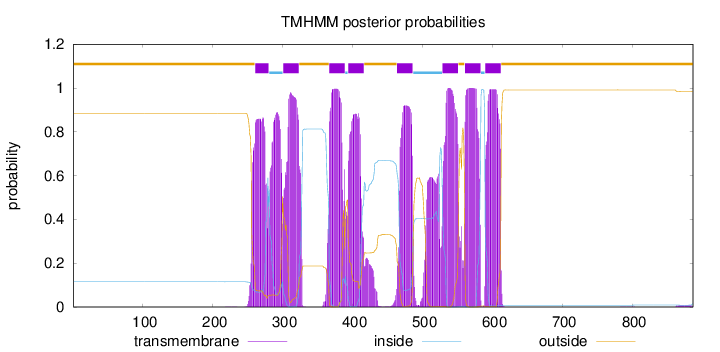
Length:
887
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
197.47948
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11506
outside
1 - 260
TMhelix
261 - 280
inside
281 - 300
TMhelix
301 - 323
outside
324 - 366
TMhelix
367 - 389
inside
390 - 393
TMhelix
394 - 416
outside
417 - 463
TMhelix
464 - 486
inside
487 - 528
TMhelix
529 - 551
outside
552 - 560
TMhelix
561 - 583
inside
584 - 589
TMhelix
590 - 612
outside
613 - 887
Population Genetic Test Statistics
Pi
380.067306
Theta
177.634021
Tajima's D
1.340145
CLR
0.291344
CSRT
0.745862706864657
Interpretation
Uncertain
