Gene
KWMTBOMO15687
Pre Gene Modal
BGIBMGA002277
Annotation
PREDICTED:_dopamine_receptor_1_[Papilio_polytes]
Full name
Dopamine receptor 1
Alternative Name
Dopamine 1-like receptor 1
Location in the cell
Extracellular Reliability : 1.741 PlasmaMembrane Reliability : 1.802
Sequence
CDS
ATGAGCATTCCTTTTCCAGATCTAGCCTTCAAGATATTGACGTGGCTGGGATACTCCAACTCGGCGTTCAACCCTATTATCTACTCGATATTCAACACGGAATTCAGGGAGGCCTTCAAGAAAATCCTCACCTCGAGGTATCCACCTTGTTGCGGCTATCAAACGGTCAGAGCGAACACGCCGACGAGAAACGACAACTTCGTCACAGATTACGGAACTAAGACATTAGTGGTACGCCGAAGCGGCTCCTTAGGTCTTTCAGGAGTAGACCCCACCCCTAGATCCTCAGCGGAATCGGTAAGACCACTACGGGAATACCACATCTGA
Protein
MSIPFPDLAFKILTWLGYSNSAFNPIIYSIFNTEFREAFKKILTSRYPPCCGYQTVRANTPTRNDNFVTDYGTKTLVVRRSGSLGLSGVDPTPRSSAESVRPLREYHI
Summary
Description
Receptor for dopamine. The activity of this receptor is mediated by G proteins which activate adenylyl cyclase. Might be involved in the processing of visual information and/or visual learning. Important for Pavlovian conditioning: required in the mushroom body as a receptor conveying unconditional stimuli information, has a role in memory formation for aversive and appetitive learning. Sleep-deprivation-induced impairments in learning can be partially explained through alterations in dopamine signaling, Dop1R1 expression levels are reduced; sleep may have a role in restoring dopamine homeostasis.
Miscellaneous
Potency of neurotransmitter agonists in stimulating cAMP production and the lack of stimulation by other transmitters and metabolites suggests this is a D1-like receptor. Low homology to vertebrate D1 receptors suggests this may be a progenitor of the D1 receptor subfamily.
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Disulfide bond
G-protein coupled receptor
Glycoprotein
Lipoprotein
Membrane
Palmitate
Phosphoprotein
Polymorphism
Receptor
Reference proteome
Signal
Transducer
Transmembrane
Transmembrane helix
Feature
chain Dopamine receptor 1
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9IYE4
A9ZLX6
A0A0K0QVU3
A0A0N1PHN8
A0A194QI65
A0A2W1BCZ2
+ More
A0A212EWQ1 U4TY40 N6UC11 N1NTX2 D6X2C4 A9NJ55 A0A1W4URH6 B4R0G6 A0A3B0KNB2 A0A0M4F308 A0A1Z2RSH3 B4K025 B4G4U7 I5ANU3 P41596-3 B4HF43 B4N9I8 B3P0M9 P41596-2 A0A0B4K6H0 P41596 B3LYB5 B4M048 T1ICS3 B4KDV5 A0A0B4KHI2 B4PTB2 A0A126GUU2 A0A1I8PSI5 A0A1I8MJ91 A0A0L0BR86 A0A067RC96 A0A2P8YCS4 A0A226DLA0 A0A1A9VG05 A0A1B0FEI1 A0A1A9YST9 A0A1B0B2Q4 A0A1A9ZYZ2 A0A1D2NMV5 A0A2S2N7W3 K7INX8 A0A232FJJ9 A0A2R7W0S5 A0A0C9S1R7 E2A2Z0 E2BH27 A0A0B5H6U8 F4X4A8 A0A158NXC2 A0A0L7QKS6 A0A151WQW0 A0A026WC04 A0A154PGY0 O44198 E9ICA7 A0A2A3ECL1 A0A3L8DZP5
A0A212EWQ1 U4TY40 N6UC11 N1NTX2 D6X2C4 A9NJ55 A0A1W4URH6 B4R0G6 A0A3B0KNB2 A0A0M4F308 A0A1Z2RSH3 B4K025 B4G4U7 I5ANU3 P41596-3 B4HF43 B4N9I8 B3P0M9 P41596-2 A0A0B4K6H0 P41596 B3LYB5 B4M048 T1ICS3 B4KDV5 A0A0B4KHI2 B4PTB2 A0A126GUU2 A0A1I8PSI5 A0A1I8MJ91 A0A0L0BR86 A0A067RC96 A0A2P8YCS4 A0A226DLA0 A0A1A9VG05 A0A1B0FEI1 A0A1A9YST9 A0A1B0B2Q4 A0A1A9ZYZ2 A0A1D2NMV5 A0A2S2N7W3 K7INX8 A0A232FJJ9 A0A2R7W0S5 A0A0C9S1R7 E2A2Z0 E2BH27 A0A0B5H6U8 F4X4A8 A0A158NXC2 A0A0L7QKS6 A0A151WQW0 A0A026WC04 A0A154PGY0 O44198 E9ICA7 A0A2A3ECL1 A0A3L8DZP5
Pubmed
19121390
18353107
26179416
26354079
28756777
22118469
+ More
23537049 18054377 18025266 18316733 20068045 21843505 23604020 18362917 19820115 17994087 15632085 7953290 10731132 12537572 8630055 7720859 12711555 17634358 18674913 12537568 12537573 12537574 16110336 17569856 17569867 17550304 25315136 26108605 24845553 29403074 27289101 20075255 28648823 20798317 21719571 21347285 24508170 9422342 21282665 30249741
23537049 18054377 18025266 18316733 20068045 21843505 23604020 18362917 19820115 17994087 15632085 7953290 10731132 12537572 8630055 7720859 12711555 17634358 18674913 12537568 12537573 12537574 16110336 17569856 17569867 17550304 25315136 26108605 24845553 29403074 27289101 20075255 28648823 20798317 21719571 21347285 24508170 9422342 21282665 30249741
EMBL
BABH01020548
AB162715
AB362162
BAF98647.1
BAG14260.1
KP784317
+ More
RSAL01000087 AKR18178.1 RVE48225.1 KQ459993 KPJ18572.1 KQ458761 KPJ05207.1 KZ150270 PZC71684.1 AGBW02011944 OWR45877.1 KB631642 ERL84903.1 APGK01029541 KB740735 ENN79190.1 BK005863 DAA64500.1 KQ971371 EFA10743.2 DQ645628 ABG29734.1 CM000364 EDX12981.1 OUUW01000013 SPP88079.1 CP012526 ALC45545.1 KX360128 ASA47151.1 CH916384 EDV91627.1 CH479179 EDW24613.1 CM000070 EIM52628.2 X77234 AE014297 BT030444 BT150144 U22106 CH480815 EDW42217.1 CH964232 EDW81664.2 CH954181 EDV48855.1 AFH06410.1 CH902617 EDV44019.2 CH940650 EDW68298.2 ACPB03020246 CH933806 EDW14952.2 AGB95944.1 CM000160 EDW97611.2 ALI30568.1 JRES01001482 KNC22547.1 KK852731 KDR17488.1 PYGN01000699 PSN42062.1 LNIX01000017 OXA45614.1 CCAG010001986 JXJN01007687 LJIJ01000002 ODN06613.1 GGMR01000611 MBY13230.1 NNAY01000127 OXU30720.1 KK854224 PTY13312.1 GBYB01014731 JAG84498.1 GL436239 EFN72198.1 GL448268 EFN84948.1 KP233833 AJF39157.1 GL888633 EGI58704.1 ADTU01000452 KQ414940 KOC59217.1 KQ982815 KYQ50244.1 KK107323 EZA52589.1 KQ434902 KZC11072.1 Y13429 CAA73841.1 GL762231 EFZ21779.1 KZ288287 PBC29430.1 QOIP01000002 RLU25877.1
RSAL01000087 AKR18178.1 RVE48225.1 KQ459993 KPJ18572.1 KQ458761 KPJ05207.1 KZ150270 PZC71684.1 AGBW02011944 OWR45877.1 KB631642 ERL84903.1 APGK01029541 KB740735 ENN79190.1 BK005863 DAA64500.1 KQ971371 EFA10743.2 DQ645628 ABG29734.1 CM000364 EDX12981.1 OUUW01000013 SPP88079.1 CP012526 ALC45545.1 KX360128 ASA47151.1 CH916384 EDV91627.1 CH479179 EDW24613.1 CM000070 EIM52628.2 X77234 AE014297 BT030444 BT150144 U22106 CH480815 EDW42217.1 CH964232 EDW81664.2 CH954181 EDV48855.1 AFH06410.1 CH902617 EDV44019.2 CH940650 EDW68298.2 ACPB03020246 CH933806 EDW14952.2 AGB95944.1 CM000160 EDW97611.2 ALI30568.1 JRES01001482 KNC22547.1 KK852731 KDR17488.1 PYGN01000699 PSN42062.1 LNIX01000017 OXA45614.1 CCAG010001986 JXJN01007687 LJIJ01000002 ODN06613.1 GGMR01000611 MBY13230.1 NNAY01000127 OXU30720.1 KK854224 PTY13312.1 GBYB01014731 JAG84498.1 GL436239 EFN72198.1 GL448268 EFN84948.1 KP233833 AJF39157.1 GL888633 EGI58704.1 ADTU01000452 KQ414940 KOC59217.1 KQ982815 KYQ50244.1 KK107323 EZA52589.1 KQ434902 KZC11072.1 Y13429 CAA73841.1 GL762231 EFZ21779.1 KZ288287 PBC29430.1 QOIP01000002 RLU25877.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000030742
+ More
UP000019118 UP000007266 UP000192221 UP000000304 UP000268350 UP000092553 UP000001070 UP000008744 UP000001819 UP000000803 UP000001292 UP000007798 UP000008711 UP000007801 UP000008792 UP000015103 UP000009192 UP000002282 UP000095300 UP000095301 UP000037069 UP000027135 UP000245037 UP000198287 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000094527 UP000002358 UP000215335 UP000000311 UP000008237 UP000007755 UP000005205 UP000053825 UP000075809 UP000053097 UP000076502 UP000242457 UP000279307
UP000019118 UP000007266 UP000192221 UP000000304 UP000268350 UP000092553 UP000001070 UP000008744 UP000001819 UP000000803 UP000001292 UP000007798 UP000008711 UP000007801 UP000008792 UP000015103 UP000009192 UP000002282 UP000095300 UP000095301 UP000037069 UP000027135 UP000245037 UP000198287 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000094527 UP000002358 UP000215335 UP000000311 UP000008237 UP000007755 UP000005205 UP000053825 UP000075809 UP000053097 UP000076502 UP000242457 UP000279307
Pfam
PF00001 7tm_1
ProteinModelPortal
H9IYE4
A9ZLX6
A0A0K0QVU3
A0A0N1PHN8
A0A194QI65
A0A2W1BCZ2
+ More
A0A212EWQ1 U4TY40 N6UC11 N1NTX2 D6X2C4 A9NJ55 A0A1W4URH6 B4R0G6 A0A3B0KNB2 A0A0M4F308 A0A1Z2RSH3 B4K025 B4G4U7 I5ANU3 P41596-3 B4HF43 B4N9I8 B3P0M9 P41596-2 A0A0B4K6H0 P41596 B3LYB5 B4M048 T1ICS3 B4KDV5 A0A0B4KHI2 B4PTB2 A0A126GUU2 A0A1I8PSI5 A0A1I8MJ91 A0A0L0BR86 A0A067RC96 A0A2P8YCS4 A0A226DLA0 A0A1A9VG05 A0A1B0FEI1 A0A1A9YST9 A0A1B0B2Q4 A0A1A9ZYZ2 A0A1D2NMV5 A0A2S2N7W3 K7INX8 A0A232FJJ9 A0A2R7W0S5 A0A0C9S1R7 E2A2Z0 E2BH27 A0A0B5H6U8 F4X4A8 A0A158NXC2 A0A0L7QKS6 A0A151WQW0 A0A026WC04 A0A154PGY0 O44198 E9ICA7 A0A2A3ECL1 A0A3L8DZP5
A0A212EWQ1 U4TY40 N6UC11 N1NTX2 D6X2C4 A9NJ55 A0A1W4URH6 B4R0G6 A0A3B0KNB2 A0A0M4F308 A0A1Z2RSH3 B4K025 B4G4U7 I5ANU3 P41596-3 B4HF43 B4N9I8 B3P0M9 P41596-2 A0A0B4K6H0 P41596 B3LYB5 B4M048 T1ICS3 B4KDV5 A0A0B4KHI2 B4PTB2 A0A126GUU2 A0A1I8PSI5 A0A1I8MJ91 A0A0L0BR86 A0A067RC96 A0A2P8YCS4 A0A226DLA0 A0A1A9VG05 A0A1B0FEI1 A0A1A9YST9 A0A1B0B2Q4 A0A1A9ZYZ2 A0A1D2NMV5 A0A2S2N7W3 K7INX8 A0A232FJJ9 A0A2R7W0S5 A0A0C9S1R7 E2A2Z0 E2BH27 A0A0B5H6U8 F4X4A8 A0A158NXC2 A0A0L7QKS6 A0A151WQW0 A0A026WC04 A0A154PGY0 O44198 E9ICA7 A0A2A3ECL1 A0A3L8DZP5
PDB
6CM4
E-value=1.67031e-08,
Score=134
Ontologies
GO
GO:0004930
GO:0016021
GO:0005886
GO:0035240
GO:0007191
GO:0001588
GO:0001965
GO:0007212
GO:0042594
GO:1990834
GO:0007612
GO:0043052
GO:0007613
GO:0009744
GO:0040040
GO:0099509
GO:0008542
GO:0008306
GO:0008355
GO:0071329
GO:0004952
GO:0005887
GO:0090328
GO:0098793
GO:0007186
GO:0004983
GO:0005634
GO:0006270
GO:0042555
GO:0003707
GO:0006281
GO:0006259
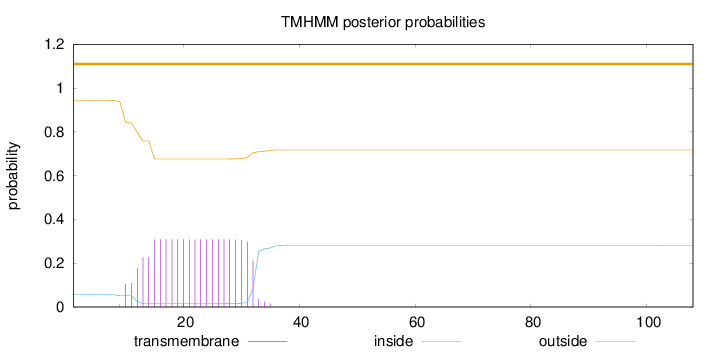
Topology
Subcellular location
Cell membrane
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.39125
Exp number, first 60 AAs:
6.3911
Total prob of N-in:
0.05611
outside
1 - 108
Population Genetic Test Statistics
Pi
130.826685
Theta
187.63539
Tajima's D
-0.94734
CLR
7.767483
CSRT
0.149892505374731
Interpretation
Uncertain
