Gene
KWMTBOMO15677
Pre Gene Modal
BGIBMGA002282
Annotation
PREDICTED:_coagulation_factor_IX-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.871
Sequence
CDS
ATGAACAATGACATTGCCATTGTCAAAGTTGACGAAGAATTCGACTTCACTCGACGAGTTAGGGGTTGTGATTTTATACCAAAGCCCGTCGCTTACAATAATTATAGTACCAATCTTGAAGAGCCAGGCGTTCGGGCCAATATCGCCGGCTGGGGTACCACCAAGCATTATGACGGTTCCGAAGGCGGGAAACCGACTCAAAACTTGCTTCAAAGCGAAGTGGAGATAATAAATAAGGACCGATGCAAAAGAAGATGGGGCAAACGTTACTACAATATCATCGATAACTACATGATCTGTTCTAAAGACAACGCGCCGGTGCTGAACGAAGTGTGTCGGGAGAAGTACGTGGATTGTACTGATATAGATTATTCGGACGAGGTTGACGGAACAAGACGTGTAATAAGTCCAGAGAAATTAGAGCTACATTCTGCTCATCACAATGTATCGGGGAGGAGAAACGTAGACAGCGGTGGATTCTGTGAGTCAGTTAAAATTGTTCTTGCTTTCAGGGTATATCTTGATTAA
Protein
MNNDIAIVKVDEEFDFTRRVRGCDFIPKPVAYNNYSTNLEEPGVRANIAGWGTTKHYDGSEGGKPTQNLLQSEVEIINKDRCKRRWGKRYYNIIDNYMICSKDNAPVLNEVCREKYVDCTDIDYSDEVDGTRRVISPEKLELHSAHHNVSGRRNVDSGGFCESVKIVLAFRVYLD
Summary
Uniprot
A0A286NZX1
H9IYE8
A0A2W1BTR5
A0A2H1VGP7
A0A3S2NDF2
A0A3S2M0D8
+ More
A0A2A4IUK6 A0A212F776 G9D4S1 G9D4S2 G9D4R0 G9D4R9 G9D4S0 A0A2W1BZ11 G9D4R2 G9D4R4 G9D4R3 G9D4R8 G9D4R7 G9D4R6 G9D4R5 D9HQ48 D9HQ88 H9IYE9 A0A0P0UWE9 A0A0N0PEI2 A0A194QJW1 H9IYE7 A0A286NZX2 A0A2W1BTQ4 A0A0N1IJE7 G9D4D7 G9D4D1 A0A0L7L8S3 G9D4D3 G9D4D6 G9D4E1 G9D4D2 G9D4E0 G9D4D8 G9D4D9 D9HQ28 D9HQ69 G9D4D4 G9D4E3 G9D4E2 H9IYF0 A0A2A4IUR8 G9D4D5 A0A0P0UWH4 A0A2W1BPQ4 A0A212F772 A0A220K8M8 A0A0L7KZA3 A0A0N0PE72 A0A2A4IWH2
A0A2A4IUK6 A0A212F776 G9D4S1 G9D4S2 G9D4R0 G9D4R9 G9D4S0 A0A2W1BZ11 G9D4R2 G9D4R4 G9D4R3 G9D4R8 G9D4R7 G9D4R6 G9D4R5 D9HQ48 D9HQ88 H9IYE9 A0A0P0UWE9 A0A0N0PEI2 A0A194QJW1 H9IYE7 A0A286NZX2 A0A2W1BTQ4 A0A0N1IJE7 G9D4D7 G9D4D1 A0A0L7L8S3 G9D4D3 G9D4D6 G9D4E1 G9D4D2 G9D4E0 G9D4D8 G9D4D9 D9HQ28 D9HQ69 G9D4D4 G9D4E3 G9D4E2 H9IYF0 A0A2A4IUR8 G9D4D5 A0A0P0UWH4 A0A2W1BPQ4 A0A212F772 A0A220K8M8 A0A0L7KZA3 A0A0N0PE72 A0A2A4IWH2
EMBL
LC316987
BBA26326.1
BABH01020509
BABH01020510
KZ149938
PZC77024.1
+ More
ODYU01002283 SOQ39562.1 RSAL01000087 RVE48232.1 RVE48233.1 NWSH01007665 PCG62872.1 AGBW02009906 OWR49591.1 JF777324 AEU11669.1 JF777325 AEU11670.1 JF777313 AEU11658.1 JF777322 AEU11667.1 JF777323 AEU11668.1 PZC77023.1 JF777315 AEU11660.1 JF777317 AEU11662.1 JF777316 AEU11661.1 JF777321 AEU11666.1 JF777320 AEU11665.1 JF777319 AEU11664.1 JF777318 AEU11663.1 HM023803 ADJ58536.1 HM023843 JF777314 ADJ58576.1 AEU11659.1 BABH01020508 LC042540 BAS69677.1 KQ459941 KPJ19183.1 KQ458761 KPJ05200.1 LC316988 BBA26327.1 PZC77025.1 KPJ19184.1 JF777190 AEU11535.1 JF777184 AEU11529.1 JTDY01002193 KOB71907.1 JF777186 AEU11531.1 JF777189 AEU11534.1 JF777194 AEU11539.1 JF777185 AEU11530.1 JF777193 AEU11538.1 JF777191 AEU11536.1 JF777192 AEU11537.1 HM023783 ADJ58516.1 HM023824 ADJ58557.1 JF777187 AEU11532.1 JF777196 AEU11541.1 JF777195 AEU11540.1 PCG62873.1 JF777188 AEU11533.1 LC042542 BAS69679.1 PZC77022.1 OWR49592.1 MF319720 ASJ26434.1 JTDY01004271 KOB68354.1 KPJ19182.1 NWSH01006308 PCG63522.1
ODYU01002283 SOQ39562.1 RSAL01000087 RVE48232.1 RVE48233.1 NWSH01007665 PCG62872.1 AGBW02009906 OWR49591.1 JF777324 AEU11669.1 JF777325 AEU11670.1 JF777313 AEU11658.1 JF777322 AEU11667.1 JF777323 AEU11668.1 PZC77023.1 JF777315 AEU11660.1 JF777317 AEU11662.1 JF777316 AEU11661.1 JF777321 AEU11666.1 JF777320 AEU11665.1 JF777319 AEU11664.1 JF777318 AEU11663.1 HM023803 ADJ58536.1 HM023843 JF777314 ADJ58576.1 AEU11659.1 BABH01020508 LC042540 BAS69677.1 KQ459941 KPJ19183.1 KQ458761 KPJ05200.1 LC316988 BBA26327.1 PZC77025.1 KPJ19184.1 JF777190 AEU11535.1 JF777184 AEU11529.1 JTDY01002193 KOB71907.1 JF777186 AEU11531.1 JF777189 AEU11534.1 JF777194 AEU11539.1 JF777185 AEU11530.1 JF777193 AEU11538.1 JF777191 AEU11536.1 JF777192 AEU11537.1 HM023783 ADJ58516.1 HM023824 ADJ58557.1 JF777187 AEU11532.1 JF777196 AEU11541.1 JF777195 AEU11540.1 PCG62873.1 JF777188 AEU11533.1 LC042542 BAS69679.1 PZC77022.1 OWR49592.1 MF319720 ASJ26434.1 JTDY01004271 KOB68354.1 KPJ19182.1 NWSH01006308 PCG63522.1
Proteomes
Pfam
PF00089 Trypsin
SUPFAM
SSF50494
SSF50494
ProteinModelPortal
A0A286NZX1
H9IYE8
A0A2W1BTR5
A0A2H1VGP7
A0A3S2NDF2
A0A3S2M0D8
+ More
A0A2A4IUK6 A0A212F776 G9D4S1 G9D4S2 G9D4R0 G9D4R9 G9D4S0 A0A2W1BZ11 G9D4R2 G9D4R4 G9D4R3 G9D4R8 G9D4R7 G9D4R6 G9D4R5 D9HQ48 D9HQ88 H9IYE9 A0A0P0UWE9 A0A0N0PEI2 A0A194QJW1 H9IYE7 A0A286NZX2 A0A2W1BTQ4 A0A0N1IJE7 G9D4D7 G9D4D1 A0A0L7L8S3 G9D4D3 G9D4D6 G9D4E1 G9D4D2 G9D4E0 G9D4D8 G9D4D9 D9HQ28 D9HQ69 G9D4D4 G9D4E3 G9D4E2 H9IYF0 A0A2A4IUR8 G9D4D5 A0A0P0UWH4 A0A2W1BPQ4 A0A212F772 A0A220K8M8 A0A0L7KZA3 A0A0N0PE72 A0A2A4IWH2
A0A2A4IUK6 A0A212F776 G9D4S1 G9D4S2 G9D4R0 G9D4R9 G9D4S0 A0A2W1BZ11 G9D4R2 G9D4R4 G9D4R3 G9D4R8 G9D4R7 G9D4R6 G9D4R5 D9HQ48 D9HQ88 H9IYE9 A0A0P0UWE9 A0A0N0PEI2 A0A194QJW1 H9IYE7 A0A286NZX2 A0A2W1BTQ4 A0A0N1IJE7 G9D4D7 G9D4D1 A0A0L7L8S3 G9D4D3 G9D4D6 G9D4E1 G9D4D2 G9D4E0 G9D4D8 G9D4D9 D9HQ28 D9HQ69 G9D4D4 G9D4E3 G9D4E2 H9IYF0 A0A2A4IUR8 G9D4D5 A0A0P0UWH4 A0A2W1BPQ4 A0A212F772 A0A220K8M8 A0A0L7KZA3 A0A0N0PE72 A0A2A4IWH2
PDB
2VU8
E-value=0.000265154,
Score=100
Ontologies
GO
Topology
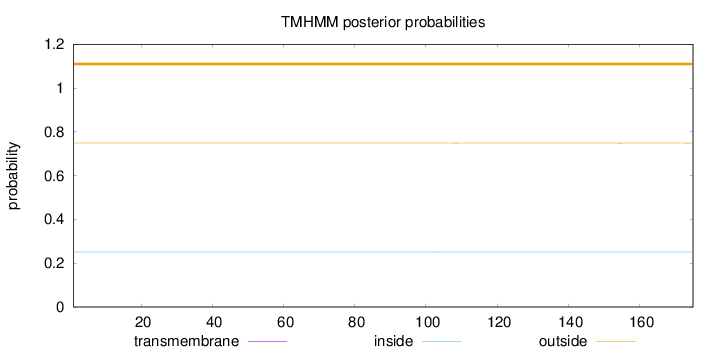
Length:
175
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00851
Exp number, first 60 AAs:
0
Total prob of N-in:
0.25135
outside
1 - 175
Population Genetic Test Statistics
Pi
212.493691
Theta
203.572451
Tajima's D
0.575774
CLR
0.024829
CSRT
0.531773411329434
Interpretation
Uncertain
