Gene
KWMTBOMO15674
Pre Gene Modal
BGIBMGA002294
Annotation
putative_BTB/POZ_domain-containing_protein_KCTD9_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 2.312
Sequence
CDS
ATGGATGTTGTCCGAGCTCTGATAAAAACATCCACATCTATTGAGCTGAGGTTTCAAGGAGTGAACTTAGCCGGAGCTGACCTGAATAGATTAGATTTGAGATTTATTAATTTTAAGTATGCCTTCTTATCTCGGTGCAATTTGAGTGGTGCTAATCTGTCTTACTGCTGTCTGGAAAGGGCGGACTTGTCGCATGCTAATTTAGAAGGTGCCCAGCTTCTTGGAGTTAAAGCTTTATGTGCTAACATGGAAGGTGCTAATTTAAAAGGTTGCAATATGGATGATCCTCACGGTAACAAAGCACTGATGGAAGGCGTTAATCTTAAAGGAGCAAATCTAGAAGGCAGTAATATGTCGGGGGTTAATTTGAGAGTGGCAACACTAAAGAACGCCAACTTACAAAACTGCGATCTGAGGACGGCTGTGCTCGCCGGTGCAGATCTGGAGTGTTGCGATCTGTCGGGCAGTGACCTCCATGAAGCTAATCTACGTGGTGCCAACCTAAAAGACGCGGCTTTCGAACTAATGCTAACTCCACTACACATGTCCCAAACTGTTAGATGA
Protein
MDVVRALIKTSTSIELRFQGVNLAGADLNRLDLRFINFKYAFLSRCNLSGANLSYCCLERADLSHANLEGAQLLGVKALCANMEGANLKGCNMDDPHGNKALMEGVNLKGANLEGSNMSGVNLRVATLKNANLQNCDLRTAVLAGADLECCDLSGSDLHEANLRGANLKDAAFELMLTPLHMSQTVR
Summary
Uniprot
H9IYG0
A0A2W1BPV7
A0A2H1VFE0
A0A2A4IWA3
A0A212F779
A0A3S2L8Q4
+ More
I4DKS6 A0A194QI15 S4PBD5 A0A0N1PJ91 A0A2P8XG49 A0A067RKL2 A0A2J7QHW9 A0A0T6B357 A0A1B6G5L4 A0A1B6MF92 A0A087TL75 A0A2L2YBS1 D6X242 A0A146L479 A0A1Y1M1S0 A0A224XPG6 T1HJ00 A0A1B6CLR5 T1JJI6 E0VTL2 K7INZ4 A0A1Q3FD83 A0A1Q3FCV8 U5EPR9 A0A131XXD9 Q176F6 B0WE08 A0A1S4FDD6 B0XJG9 A0A182H7L8 E2B572 A0A182H2K3 A0A147BQN7 A0A154P877 A0A2R7WVF9 A0A232F1J6 B4JY58 B4LWA4 A0A131XCP7 L7LWQ0 A0A310STI7 B4NID0 A0A026W5K6 A0A3L8E390 A0A224YIT9 A0A131YY51 A0A1E1X2K6 A0A3S3NVW8 A0A3L8D421 F4WDS6 A0A151HZS3 A0A1I8QB66 A0A0L0CL00 A0A1I8M292 A0A1W4WB27 B3LY16 A0A151JAC7 A0A1A9WZ25 A0A158NNG2 A0A0K8TNB4 B4K8Z5 A0A0M9A3M6 A0A1B0AJX6 W8BXN1 A0A034WQZ4 A0A1B0FP36 A0A1A9UWY4 A0A1B0D3U6 Q295F9 B4GDY1 A0A0A1WUD5 A0A1L8DAW8 A0A0K8UCA2 B3P204 B4PTS3 A0A0A1WUV3 A0A1A9XUC3 A0A0C9S1Z1 B4I3N6 A0A151JWL8 B4QVH5 B9A0N7 Q8T0F7 A0A3B0K4M9 A0A195CA13 E9FVE8 A0A0L7R2F3 A0A0N8ASY2 A0A1B6K5M0 A0A2R5LEY6 A0A088AE12 A0A2A3ESP7 A0A0P4WNI0 A0A3R7PFD9
I4DKS6 A0A194QI15 S4PBD5 A0A0N1PJ91 A0A2P8XG49 A0A067RKL2 A0A2J7QHW9 A0A0T6B357 A0A1B6G5L4 A0A1B6MF92 A0A087TL75 A0A2L2YBS1 D6X242 A0A146L479 A0A1Y1M1S0 A0A224XPG6 T1HJ00 A0A1B6CLR5 T1JJI6 E0VTL2 K7INZ4 A0A1Q3FD83 A0A1Q3FCV8 U5EPR9 A0A131XXD9 Q176F6 B0WE08 A0A1S4FDD6 B0XJG9 A0A182H7L8 E2B572 A0A182H2K3 A0A147BQN7 A0A154P877 A0A2R7WVF9 A0A232F1J6 B4JY58 B4LWA4 A0A131XCP7 L7LWQ0 A0A310STI7 B4NID0 A0A026W5K6 A0A3L8E390 A0A224YIT9 A0A131YY51 A0A1E1X2K6 A0A3S3NVW8 A0A3L8D421 F4WDS6 A0A151HZS3 A0A1I8QB66 A0A0L0CL00 A0A1I8M292 A0A1W4WB27 B3LY16 A0A151JAC7 A0A1A9WZ25 A0A158NNG2 A0A0K8TNB4 B4K8Z5 A0A0M9A3M6 A0A1B0AJX6 W8BXN1 A0A034WQZ4 A0A1B0FP36 A0A1A9UWY4 A0A1B0D3U6 Q295F9 B4GDY1 A0A0A1WUD5 A0A1L8DAW8 A0A0K8UCA2 B3P204 B4PTS3 A0A0A1WUV3 A0A1A9XUC3 A0A0C9S1Z1 B4I3N6 A0A151JWL8 B4QVH5 B9A0N7 Q8T0F7 A0A3B0K4M9 A0A195CA13 E9FVE8 A0A0L7R2F3 A0A0N8ASY2 A0A1B6K5M0 A0A2R5LEY6 A0A088AE12 A0A2A3ESP7 A0A0P4WNI0 A0A3R7PFD9
Pubmed
19121390
28756777
22118469
22651552
26354079
23622113
+ More
29403074 24845553 26561354 18362917 19820115 26823975 28004739 20566863 20075255 17510324 26483478 20798317 29652888 28648823 17994087 28049606 25576852 24508170 30249741 28797301 26830274 28503490 21719571 26108605 25315136 21347285 26369729 24495485 25348373 15632085 25830018 17550304 26131772 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21292972
29403074 24845553 26561354 18362917 19820115 26823975 28004739 20566863 20075255 17510324 26483478 20798317 29652888 28648823 17994087 28049606 25576852 24508170 30249741 28797301 26830274 28503490 21719571 26108605 25315136 21347285 26369729 24495485 25348373 15632085 25830018 17550304 26131772 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21292972
EMBL
BABH01020501
KZ149938
PZC77018.1
ODYU01002283
SOQ39558.1
NWSH01006469
+ More
PCG63410.1 AGBW02009906 OWR49595.1 RSAL01000087 RVE48238.1 AK401894 BAM18516.1 KQ458761 KPJ05198.1 GAIX01005962 JAA86598.1 KQ459941 KPJ19180.1 PYGN01002270 PSN30952.1 KK852458 KDR23553.1 NEVH01013964 PNF28166.1 LJIG01016044 KRT81784.1 GECZ01012034 JAS57735.1 GEBQ01005418 JAT34559.1 KK115731 KFM65864.1 IAAA01012894 IAAA01012895 LAA05447.1 KQ971371 EFA10211.2 GDHC01015930 JAQ02699.1 GEZM01044369 JAV78275.1 GFTR01005974 JAW10452.1 ACPB03019207 GEDC01023067 JAS14231.1 JH432222 DS235767 EEB16739.1 GFDL01009562 JAV25483.1 GFDL01009712 JAV25333.1 GANO01003605 JAB56266.1 GEFM01004459 JAP71337.1 CH477387 EAT42060.1 DS231903 EDS45138.1 DS233515 EDS30372.1 JXUM01116570 KQ565998 KXJ70463.1 GL445725 EFN89166.1 JXUM01105536 KQ564974 KXJ71480.1 GEGO01002327 JAR93077.1 KQ434839 KZC08077.1 KK855690 PTY23547.1 NNAY01001318 OXU24380.1 CH916377 EDV90620.1 CH940650 EDW67638.2 GEFH01005105 JAP63476.1 GACK01009007 JAA56027.1 KQ760539 OAD59993.1 CH964272 EDW83712.1 KK107432 EZA50896.1 QOIP01000001 RLU26855.1 GFPF01006421 MAA17567.1 GEDV01004428 JAP84129.1 GFAC01005730 JAT93458.1 NCKU01007356 NCKU01007123 RWS02694.1 RWS02876.1 QOIP01000013 RLU15150.1 GL888092 EGI67636.1 KQ976689 KYM78121.1 JRES01000338 KNC32149.1 CH902617 EDV42872.1 KQ979332 KYN21903.1 ADTU01021487 ADTU01021488 ADTU01021489 ADTU01021490 GDAI01001721 JAI15882.1 CH933806 EDW16592.1 KQ435758 KOX75776.1 GAMC01002488 JAC04068.1 GAKP01001888 JAC57064.1 CCAG010007645 AJVK01023707 CM000070 EAL28753.1 CH479182 EDW34642.1 GBXI01012162 JAD02130.1 GFDF01010577 JAV03507.1 GDHF01028037 JAI24277.1 CH954181 EDV47777.1 CM000160 EDW95656.1 GBXI01011881 JAD02411.1 GBZX01001806 JAG90934.1 CH480821 EDW54829.1 KQ981632 KYN38924.1 CM000364 EDX11600.1 AE014297 AAF52131.5 AY069350 AAL39495.1 OUUW01000014 SPP88253.1 KQ978068 KYM97642.1 GL732525 EFX89116.1 KQ414666 KOC65057.1 GDIQ01246003 JAK05722.1 GECU01000956 JAT06751.1 GGLE01003968 MBY08094.1 KZ288187 PBC34727.1 GDRN01076296 GDRN01076295 JAI62916.1 QCYY01003225 ROT64595.1
PCG63410.1 AGBW02009906 OWR49595.1 RSAL01000087 RVE48238.1 AK401894 BAM18516.1 KQ458761 KPJ05198.1 GAIX01005962 JAA86598.1 KQ459941 KPJ19180.1 PYGN01002270 PSN30952.1 KK852458 KDR23553.1 NEVH01013964 PNF28166.1 LJIG01016044 KRT81784.1 GECZ01012034 JAS57735.1 GEBQ01005418 JAT34559.1 KK115731 KFM65864.1 IAAA01012894 IAAA01012895 LAA05447.1 KQ971371 EFA10211.2 GDHC01015930 JAQ02699.1 GEZM01044369 JAV78275.1 GFTR01005974 JAW10452.1 ACPB03019207 GEDC01023067 JAS14231.1 JH432222 DS235767 EEB16739.1 GFDL01009562 JAV25483.1 GFDL01009712 JAV25333.1 GANO01003605 JAB56266.1 GEFM01004459 JAP71337.1 CH477387 EAT42060.1 DS231903 EDS45138.1 DS233515 EDS30372.1 JXUM01116570 KQ565998 KXJ70463.1 GL445725 EFN89166.1 JXUM01105536 KQ564974 KXJ71480.1 GEGO01002327 JAR93077.1 KQ434839 KZC08077.1 KK855690 PTY23547.1 NNAY01001318 OXU24380.1 CH916377 EDV90620.1 CH940650 EDW67638.2 GEFH01005105 JAP63476.1 GACK01009007 JAA56027.1 KQ760539 OAD59993.1 CH964272 EDW83712.1 KK107432 EZA50896.1 QOIP01000001 RLU26855.1 GFPF01006421 MAA17567.1 GEDV01004428 JAP84129.1 GFAC01005730 JAT93458.1 NCKU01007356 NCKU01007123 RWS02694.1 RWS02876.1 QOIP01000013 RLU15150.1 GL888092 EGI67636.1 KQ976689 KYM78121.1 JRES01000338 KNC32149.1 CH902617 EDV42872.1 KQ979332 KYN21903.1 ADTU01021487 ADTU01021488 ADTU01021489 ADTU01021490 GDAI01001721 JAI15882.1 CH933806 EDW16592.1 KQ435758 KOX75776.1 GAMC01002488 JAC04068.1 GAKP01001888 JAC57064.1 CCAG010007645 AJVK01023707 CM000070 EAL28753.1 CH479182 EDW34642.1 GBXI01012162 JAD02130.1 GFDF01010577 JAV03507.1 GDHF01028037 JAI24277.1 CH954181 EDV47777.1 CM000160 EDW95656.1 GBXI01011881 JAD02411.1 GBZX01001806 JAG90934.1 CH480821 EDW54829.1 KQ981632 KYN38924.1 CM000364 EDX11600.1 AE014297 AAF52131.5 AY069350 AAL39495.1 OUUW01000014 SPP88253.1 KQ978068 KYM97642.1 GL732525 EFX89116.1 KQ414666 KOC65057.1 GDIQ01246003 JAK05722.1 GECU01000956 JAT06751.1 GGLE01003968 MBY08094.1 KZ288187 PBC34727.1 GDRN01076296 GDRN01076295 JAI62916.1 QCYY01003225 ROT64595.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000245037 UP000027135 UP000235965 UP000054359 UP000007266 UP000015103 UP000009046 UP000002358 UP000008820 UP000002320 UP000069940 UP000249989 UP000008237 UP000076502 UP000215335 UP000001070 UP000008792 UP000007798 UP000053097 UP000279307 UP000285301 UP000007755 UP000078540 UP000095300 UP000037069 UP000095301 UP000192223 UP000007801 UP000078492 UP000091820 UP000005205 UP000009192 UP000053105 UP000092445 UP000092444 UP000078200 UP000092462 UP000001819 UP000008744 UP000008711 UP000002282 UP000092443 UP000001292 UP000078541 UP000000304 UP000000803 UP000268350 UP000078542 UP000000305 UP000053825 UP000005203 UP000242457 UP000283509
UP000245037 UP000027135 UP000235965 UP000054359 UP000007266 UP000015103 UP000009046 UP000002358 UP000008820 UP000002320 UP000069940 UP000249989 UP000008237 UP000076502 UP000215335 UP000001070 UP000008792 UP000007798 UP000053097 UP000279307 UP000285301 UP000007755 UP000078540 UP000095300 UP000037069 UP000095301 UP000192223 UP000007801 UP000078492 UP000091820 UP000005205 UP000009192 UP000053105 UP000092445 UP000092444 UP000078200 UP000092462 UP000001819 UP000008744 UP000008711 UP000002282 UP000092443 UP000001292 UP000078541 UP000000304 UP000000803 UP000268350 UP000078542 UP000000305 UP000053825 UP000005203 UP000242457 UP000283509
Interpro
ProteinModelPortal
H9IYG0
A0A2W1BPV7
A0A2H1VFE0
A0A2A4IWA3
A0A212F779
A0A3S2L8Q4
+ More
I4DKS6 A0A194QI15 S4PBD5 A0A0N1PJ91 A0A2P8XG49 A0A067RKL2 A0A2J7QHW9 A0A0T6B357 A0A1B6G5L4 A0A1B6MF92 A0A087TL75 A0A2L2YBS1 D6X242 A0A146L479 A0A1Y1M1S0 A0A224XPG6 T1HJ00 A0A1B6CLR5 T1JJI6 E0VTL2 K7INZ4 A0A1Q3FD83 A0A1Q3FCV8 U5EPR9 A0A131XXD9 Q176F6 B0WE08 A0A1S4FDD6 B0XJG9 A0A182H7L8 E2B572 A0A182H2K3 A0A147BQN7 A0A154P877 A0A2R7WVF9 A0A232F1J6 B4JY58 B4LWA4 A0A131XCP7 L7LWQ0 A0A310STI7 B4NID0 A0A026W5K6 A0A3L8E390 A0A224YIT9 A0A131YY51 A0A1E1X2K6 A0A3S3NVW8 A0A3L8D421 F4WDS6 A0A151HZS3 A0A1I8QB66 A0A0L0CL00 A0A1I8M292 A0A1W4WB27 B3LY16 A0A151JAC7 A0A1A9WZ25 A0A158NNG2 A0A0K8TNB4 B4K8Z5 A0A0M9A3M6 A0A1B0AJX6 W8BXN1 A0A034WQZ4 A0A1B0FP36 A0A1A9UWY4 A0A1B0D3U6 Q295F9 B4GDY1 A0A0A1WUD5 A0A1L8DAW8 A0A0K8UCA2 B3P204 B4PTS3 A0A0A1WUV3 A0A1A9XUC3 A0A0C9S1Z1 B4I3N6 A0A151JWL8 B4QVH5 B9A0N7 Q8T0F7 A0A3B0K4M9 A0A195CA13 E9FVE8 A0A0L7R2F3 A0A0N8ASY2 A0A1B6K5M0 A0A2R5LEY6 A0A088AE12 A0A2A3ESP7 A0A0P4WNI0 A0A3R7PFD9
I4DKS6 A0A194QI15 S4PBD5 A0A0N1PJ91 A0A2P8XG49 A0A067RKL2 A0A2J7QHW9 A0A0T6B357 A0A1B6G5L4 A0A1B6MF92 A0A087TL75 A0A2L2YBS1 D6X242 A0A146L479 A0A1Y1M1S0 A0A224XPG6 T1HJ00 A0A1B6CLR5 T1JJI6 E0VTL2 K7INZ4 A0A1Q3FD83 A0A1Q3FCV8 U5EPR9 A0A131XXD9 Q176F6 B0WE08 A0A1S4FDD6 B0XJG9 A0A182H7L8 E2B572 A0A182H2K3 A0A147BQN7 A0A154P877 A0A2R7WVF9 A0A232F1J6 B4JY58 B4LWA4 A0A131XCP7 L7LWQ0 A0A310STI7 B4NID0 A0A026W5K6 A0A3L8E390 A0A224YIT9 A0A131YY51 A0A1E1X2K6 A0A3S3NVW8 A0A3L8D421 F4WDS6 A0A151HZS3 A0A1I8QB66 A0A0L0CL00 A0A1I8M292 A0A1W4WB27 B3LY16 A0A151JAC7 A0A1A9WZ25 A0A158NNG2 A0A0K8TNB4 B4K8Z5 A0A0M9A3M6 A0A1B0AJX6 W8BXN1 A0A034WQZ4 A0A1B0FP36 A0A1A9UWY4 A0A1B0D3U6 Q295F9 B4GDY1 A0A0A1WUD5 A0A1L8DAW8 A0A0K8UCA2 B3P204 B4PTS3 A0A0A1WUV3 A0A1A9XUC3 A0A0C9S1Z1 B4I3N6 A0A151JWL8 B4QVH5 B9A0N7 Q8T0F7 A0A3B0K4M9 A0A195CA13 E9FVE8 A0A0L7R2F3 A0A0N8ASY2 A0A1B6K5M0 A0A2R5LEY6 A0A088AE12 A0A2A3ESP7 A0A0P4WNI0 A0A3R7PFD9
PDB
5DZB
E-value=1.6529e-17,
Score=215
Ontologies
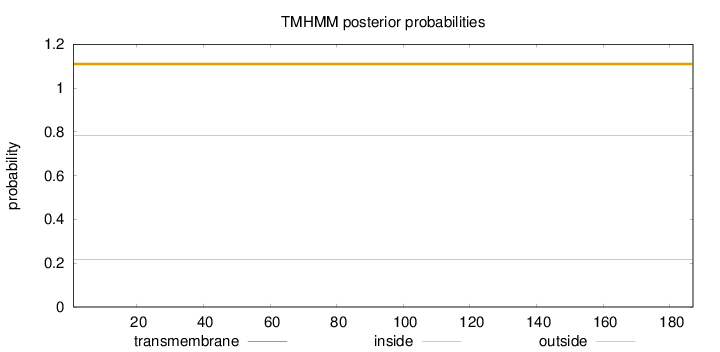
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00317
Exp number, first 60 AAs:
0.0029
Total prob of N-in:
0.21786
outside
1 - 187
Population Genetic Test Statistics
Pi
331.727196
Theta
175.87276
Tajima's D
0.7804
CLR
163.161118
CSRT
0.595320233988301
Interpretation
Uncertain
