Gene
KWMTBOMO15673
Pre Gene Modal
BGIBMGA002294
Annotation
PREDICTED:_BTB/POZ_domain-containing_protein_KCTD9_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.635 PlasmaMembrane Reliability : 1.211
Sequence
CDS
ATGAAACGTGCATTCGTCTACTTAAGTCATAAGAAACAGAATGGCAAAGTTTTGGTTGTGGAAGACACGCTTGAAGCCTTTAAATTGGAAATATCAAATGTTTTTGAAATAGGCGGAGATTTTCGCTTATTCACTGAAAATGGTTGTGAGATTGTTGACGTTCGAGTTATAAGAGACGACGAGAAGCTGTATGTCAACTGCGGCGATAGCAAAAGCTGTGATACTATTGAAAGTTCCTTGGCAAAATTATCATTGAATAGGTGTTCCGAAGTATCGGATTGGATAACACTAAATGTGGGGGGACGTTATTTTACTACTTCTCGCTCAACATTACAGTCTAAAGAGCCACTTTCGATGCTGGCTAGAATGTTTGCTGATGATAATAATATGTATTTAATGAATCCAAGTGCCACAGACTCGAGGGGAGCGTACTTAATAGATAGAAGCCCAGAGTATTTTGAACCTATTCTAAACTATTTGAGACATGGAGAAGTTATAATAGATAAATATGTAAACCCTAGAGGTGTTCTAGAAGAGGCTGTATTTTATGGTAAACTAATACTCAAAGAGACTAATTTCAAACAATACAATGGTTTCAAATATAATTACATAGTTACATTATATACACATATATTTATGTTACACTATAAGCACCCTCTTCCAATCCTTGCCAATCTGATTAAAGCCTGTGATAAGAGCATTGTCATATCATAG
Protein
MKRAFVYLSHKKQNGKVLVVEDTLEAFKLEISNVFEIGGDFRLFTENGCEIVDVRVIRDDEKLYVNCGDSKSCDTIESSLAKLSLNRCSEVSDWITLNVGGRYFTTSRSTLQSKEPLSMLARMFADDNNMYLMNPSATDSRGAYLIDRSPEYFEPILNYLRHGEVIIDKYVNPRGVLEEAVFYGKLILKETNFKQYNGFKYNYIVTLYTHIFMLHYKHPLPILANLIKACDKSIVIS
Summary
Uniprot
H9IYG0
A0A2H1VFE0
A0A2A4IWA3
A0A2W1BPV7
A0A0N1PJ91
A0A194QI15
+ More
A0A3S2L8Q4 A0A0L7L5F6 A0A212F779 A0A1B6K5M0 A0A1B6G5L4 A0A1B6MF92 A0A224XPG6 A0A2J7QHW9 E1ZWR2 A0A067RKL2 A0A026VUV7 A0A151JWL8 A0A3L8D421 A0A026W5K6 A0A3L8E390 A0A2R7WVF9 A0A0C9RT80 K7INZ4 A0A224YIT9 A0A131YY51 A0A0C9R6G4 A0A195CA13 A0A2R5LEY6 A0A1Y1M1S0 A0A232F1J6 A0A2A3ESP7 A0A2P8XG49 A0A310STI7 A0A151HZS3 A0A158NNG2 A0A088AE12 A0A3S3NVW8 A0A3Q1EV98 E2B572 A0A151JAC7 A0A3Q1CUE0 A0A3Q1CV23 A0A3P8U2Z0 T1HJ00 A0A0S7GWX7 A0A1A7X1N4 A0A0M9A3M6 T1JJI6 A0A087TL75 A0A147BQN7 A0A154P877 A0A0P5K3F1 A0A1A8U3S0 A0A1A8MCC2 A0A1A8D6A3 A0A1A8I6Y7 A0A1A8JLT0 A0A1A8PXN1 A0A1A8LDX9 A0A3B3DQ97 H3BBI3 A0A1A8VJG5 A0A1A8NLV4 A0A1A8FDN3 A0A1A8BK93 E9FVE8 A0A0P4XQ40 A0A1A8AE14 A0A1A8S7Y5 A0A3B1JRV5 A0A1A8Q9I1 A0A3B4BMT1 A0A3Q3K2L4 H3D1P9 A0A3B4EAE7 A0A0P6DYW3 H2UZ09 A0A1A8J1I8 A0A3B1JT66 W5U9E7 A0A1A8UMS9 A0A2L2YBS1 A0A0N8ASY2 A0A0P5LCU7 A0A1A8PCB0 A0A1A8KCG1 A0A1A8MU89 A0A1A8CNE5 A0A1A8GLA4 A0A0P4ZGI8 A0A1S3HM79 A0A3B4EV49 A0A3Q2VKF6 A0A3P8Q3U5 A0A3P9BKA9 A0A3Q1IY29
A0A3S2L8Q4 A0A0L7L5F6 A0A212F779 A0A1B6K5M0 A0A1B6G5L4 A0A1B6MF92 A0A224XPG6 A0A2J7QHW9 E1ZWR2 A0A067RKL2 A0A026VUV7 A0A151JWL8 A0A3L8D421 A0A026W5K6 A0A3L8E390 A0A2R7WVF9 A0A0C9RT80 K7INZ4 A0A224YIT9 A0A131YY51 A0A0C9R6G4 A0A195CA13 A0A2R5LEY6 A0A1Y1M1S0 A0A232F1J6 A0A2A3ESP7 A0A2P8XG49 A0A310STI7 A0A151HZS3 A0A158NNG2 A0A088AE12 A0A3S3NVW8 A0A3Q1EV98 E2B572 A0A151JAC7 A0A3Q1CUE0 A0A3Q1CV23 A0A3P8U2Z0 T1HJ00 A0A0S7GWX7 A0A1A7X1N4 A0A0M9A3M6 T1JJI6 A0A087TL75 A0A147BQN7 A0A154P877 A0A0P5K3F1 A0A1A8U3S0 A0A1A8MCC2 A0A1A8D6A3 A0A1A8I6Y7 A0A1A8JLT0 A0A1A8PXN1 A0A1A8LDX9 A0A3B3DQ97 H3BBI3 A0A1A8VJG5 A0A1A8NLV4 A0A1A8FDN3 A0A1A8BK93 E9FVE8 A0A0P4XQ40 A0A1A8AE14 A0A1A8S7Y5 A0A3B1JRV5 A0A1A8Q9I1 A0A3B4BMT1 A0A3Q3K2L4 H3D1P9 A0A3B4EAE7 A0A0P6DYW3 H2UZ09 A0A1A8J1I8 A0A3B1JT66 W5U9E7 A0A1A8UMS9 A0A2L2YBS1 A0A0N8ASY2 A0A0P5LCU7 A0A1A8PCB0 A0A1A8KCG1 A0A1A8MU89 A0A1A8CNE5 A0A1A8GLA4 A0A0P4ZGI8 A0A1S3HM79 A0A3B4EV49 A0A3Q2VKF6 A0A3P8Q3U5 A0A3P9BKA9 A0A3Q1IY29
Pubmed
EMBL
BABH01020501
ODYU01002283
SOQ39558.1
NWSH01006469
PCG63410.1
KZ149938
+ More
PZC77018.1 KQ459941 KPJ19180.1 KQ458761 KPJ05198.1 RSAL01000087 RVE48238.1 JTDY01002810 KOB70682.1 AGBW02009906 OWR49595.1 GECU01000956 JAT06751.1 GECZ01012034 JAS57735.1 GEBQ01005418 JAT34559.1 GFTR01005974 JAW10452.1 NEVH01013964 PNF28166.1 GL434893 EFN74377.1 KK852458 KDR23553.1 KK107804 EZA47573.1 KQ981632 KYN38924.1 QOIP01000013 RLU15150.1 KK107432 EZA50896.1 QOIP01000001 RLU26855.1 KK855690 PTY23547.1 GBYB01011915 GBYB01011917 JAG81682.1 JAG81684.1 GFPF01006421 MAA17567.1 GEDV01004428 JAP84129.1 GBYB01011919 JAG81686.1 KQ978068 KYM97642.1 GGLE01003968 MBY08094.1 GEZM01044369 JAV78275.1 NNAY01001318 OXU24380.1 KZ288187 PBC34727.1 PYGN01002270 PSN30952.1 KQ760539 OAD59993.1 KQ976689 KYM78121.1 ADTU01021487 ADTU01021488 ADTU01021489 ADTU01021490 NCKU01007356 NCKU01007123 RWS02694.1 RWS02876.1 GL445725 EFN89166.1 KQ979332 KYN21903.1 ACPB03019207 GBYX01448179 JAO33295.1 HADW01010420 HADX01006123 SBP11820.1 KQ435758 KOX75776.1 JH432222 KK115731 KFM65864.1 GEGO01002327 JAR93077.1 KQ434839 KZC08077.1 GDIQ01191519 JAK60206.1 HADY01020827 HAEJ01002492 SBS42949.1 HAEF01013421 SBR54580.1 HAEA01000383 SBQ28863.1 HAED01006605 SBQ92706.1 HAEE01000867 SBR20883.1 HAEH01008991 SBR85983.1 HAEF01005673 HAEG01006444 SBR43055.1 AFYH01036866 AFYH01036867 HAEJ01019215 SBS59672.1 HAEI01000054 SBR69988.1 HAEB01009712 HAEC01014072 SBQ56239.1 HADZ01003367 HAEA01015055 SBP67308.1 GL732525 EFX89116.1 GDIP01238146 JAI85255.1 HADY01014839 SBP53324.1 HAEH01022262 SBS13664.1 HAEH01010569 SBR90048.1 GDIQ01070223 JAN24514.1 HAED01017070 SBR03515.1 JT407950 JT415405 AHH38184.1 HAEJ01007946 SBS48403.1 IAAA01012894 IAAA01012895 LAA05447.1 GDIQ01246003 JAK05722.1 GDIQ01171584 JAK80141.1 HAEH01006362 HAEI01010682 SBR79025.1 HAED01001413 HAEE01009846 SBR29896.1 HAEF01009703 HAEF01018992 HAEG01016547 SBR60151.1 HADZ01016334 HAEA01004438 SBP80275.1 HAEB01002233 HAEB01020030 HAEC01004533 SBQ72610.1 GDIP01214196 JAJ09206.1
PZC77018.1 KQ459941 KPJ19180.1 KQ458761 KPJ05198.1 RSAL01000087 RVE48238.1 JTDY01002810 KOB70682.1 AGBW02009906 OWR49595.1 GECU01000956 JAT06751.1 GECZ01012034 JAS57735.1 GEBQ01005418 JAT34559.1 GFTR01005974 JAW10452.1 NEVH01013964 PNF28166.1 GL434893 EFN74377.1 KK852458 KDR23553.1 KK107804 EZA47573.1 KQ981632 KYN38924.1 QOIP01000013 RLU15150.1 KK107432 EZA50896.1 QOIP01000001 RLU26855.1 KK855690 PTY23547.1 GBYB01011915 GBYB01011917 JAG81682.1 JAG81684.1 GFPF01006421 MAA17567.1 GEDV01004428 JAP84129.1 GBYB01011919 JAG81686.1 KQ978068 KYM97642.1 GGLE01003968 MBY08094.1 GEZM01044369 JAV78275.1 NNAY01001318 OXU24380.1 KZ288187 PBC34727.1 PYGN01002270 PSN30952.1 KQ760539 OAD59993.1 KQ976689 KYM78121.1 ADTU01021487 ADTU01021488 ADTU01021489 ADTU01021490 NCKU01007356 NCKU01007123 RWS02694.1 RWS02876.1 GL445725 EFN89166.1 KQ979332 KYN21903.1 ACPB03019207 GBYX01448179 JAO33295.1 HADW01010420 HADX01006123 SBP11820.1 KQ435758 KOX75776.1 JH432222 KK115731 KFM65864.1 GEGO01002327 JAR93077.1 KQ434839 KZC08077.1 GDIQ01191519 JAK60206.1 HADY01020827 HAEJ01002492 SBS42949.1 HAEF01013421 SBR54580.1 HAEA01000383 SBQ28863.1 HAED01006605 SBQ92706.1 HAEE01000867 SBR20883.1 HAEH01008991 SBR85983.1 HAEF01005673 HAEG01006444 SBR43055.1 AFYH01036866 AFYH01036867 HAEJ01019215 SBS59672.1 HAEI01000054 SBR69988.1 HAEB01009712 HAEC01014072 SBQ56239.1 HADZ01003367 HAEA01015055 SBP67308.1 GL732525 EFX89116.1 GDIP01238146 JAI85255.1 HADY01014839 SBP53324.1 HAEH01022262 SBS13664.1 HAEH01010569 SBR90048.1 GDIQ01070223 JAN24514.1 HAED01017070 SBR03515.1 JT407950 JT415405 AHH38184.1 HAEJ01007946 SBS48403.1 IAAA01012894 IAAA01012895 LAA05447.1 GDIQ01246003 JAK05722.1 GDIQ01171584 JAK80141.1 HAEH01006362 HAEI01010682 SBR79025.1 HAED01001413 HAEE01009846 SBR29896.1 HAEF01009703 HAEF01018992 HAEG01016547 SBR60151.1 HADZ01016334 HAEA01004438 SBP80275.1 HAEB01002233 HAEB01020030 HAEC01004533 SBQ72610.1 GDIP01214196 JAJ09206.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000037510
+ More
UP000007151 UP000235965 UP000000311 UP000027135 UP000053097 UP000078541 UP000279307 UP000002358 UP000078542 UP000215335 UP000242457 UP000245037 UP000078540 UP000005205 UP000005203 UP000285301 UP000257200 UP000008237 UP000078492 UP000257160 UP000265080 UP000015103 UP000053105 UP000054359 UP000076502 UP000261560 UP000008672 UP000000305 UP000018467 UP000261520 UP000261600 UP000007303 UP000261440 UP000005226 UP000221080 UP000085678 UP000261460 UP000264840 UP000265100 UP000265160 UP000265040
UP000007151 UP000235965 UP000000311 UP000027135 UP000053097 UP000078541 UP000279307 UP000002358 UP000078542 UP000215335 UP000242457 UP000245037 UP000078540 UP000005205 UP000005203 UP000285301 UP000257200 UP000008237 UP000078492 UP000257160 UP000265080 UP000015103 UP000053105 UP000054359 UP000076502 UP000261560 UP000008672 UP000000305 UP000018467 UP000261520 UP000261600 UP000007303 UP000261440 UP000005226 UP000221080 UP000085678 UP000261460 UP000264840 UP000265100 UP000265160 UP000265040
PRIDE
Interpro
ProteinModelPortal
H9IYG0
A0A2H1VFE0
A0A2A4IWA3
A0A2W1BPV7
A0A0N1PJ91
A0A194QI15
+ More
A0A3S2L8Q4 A0A0L7L5F6 A0A212F779 A0A1B6K5M0 A0A1B6G5L4 A0A1B6MF92 A0A224XPG6 A0A2J7QHW9 E1ZWR2 A0A067RKL2 A0A026VUV7 A0A151JWL8 A0A3L8D421 A0A026W5K6 A0A3L8E390 A0A2R7WVF9 A0A0C9RT80 K7INZ4 A0A224YIT9 A0A131YY51 A0A0C9R6G4 A0A195CA13 A0A2R5LEY6 A0A1Y1M1S0 A0A232F1J6 A0A2A3ESP7 A0A2P8XG49 A0A310STI7 A0A151HZS3 A0A158NNG2 A0A088AE12 A0A3S3NVW8 A0A3Q1EV98 E2B572 A0A151JAC7 A0A3Q1CUE0 A0A3Q1CV23 A0A3P8U2Z0 T1HJ00 A0A0S7GWX7 A0A1A7X1N4 A0A0M9A3M6 T1JJI6 A0A087TL75 A0A147BQN7 A0A154P877 A0A0P5K3F1 A0A1A8U3S0 A0A1A8MCC2 A0A1A8D6A3 A0A1A8I6Y7 A0A1A8JLT0 A0A1A8PXN1 A0A1A8LDX9 A0A3B3DQ97 H3BBI3 A0A1A8VJG5 A0A1A8NLV4 A0A1A8FDN3 A0A1A8BK93 E9FVE8 A0A0P4XQ40 A0A1A8AE14 A0A1A8S7Y5 A0A3B1JRV5 A0A1A8Q9I1 A0A3B4BMT1 A0A3Q3K2L4 H3D1P9 A0A3B4EAE7 A0A0P6DYW3 H2UZ09 A0A1A8J1I8 A0A3B1JT66 W5U9E7 A0A1A8UMS9 A0A2L2YBS1 A0A0N8ASY2 A0A0P5LCU7 A0A1A8PCB0 A0A1A8KCG1 A0A1A8MU89 A0A1A8CNE5 A0A1A8GLA4 A0A0P4ZGI8 A0A1S3HM79 A0A3B4EV49 A0A3Q2VKF6 A0A3P8Q3U5 A0A3P9BKA9 A0A3Q1IY29
A0A3S2L8Q4 A0A0L7L5F6 A0A212F779 A0A1B6K5M0 A0A1B6G5L4 A0A1B6MF92 A0A224XPG6 A0A2J7QHW9 E1ZWR2 A0A067RKL2 A0A026VUV7 A0A151JWL8 A0A3L8D421 A0A026W5K6 A0A3L8E390 A0A2R7WVF9 A0A0C9RT80 K7INZ4 A0A224YIT9 A0A131YY51 A0A0C9R6G4 A0A195CA13 A0A2R5LEY6 A0A1Y1M1S0 A0A232F1J6 A0A2A3ESP7 A0A2P8XG49 A0A310STI7 A0A151HZS3 A0A158NNG2 A0A088AE12 A0A3S3NVW8 A0A3Q1EV98 E2B572 A0A151JAC7 A0A3Q1CUE0 A0A3Q1CV23 A0A3P8U2Z0 T1HJ00 A0A0S7GWX7 A0A1A7X1N4 A0A0M9A3M6 T1JJI6 A0A087TL75 A0A147BQN7 A0A154P877 A0A0P5K3F1 A0A1A8U3S0 A0A1A8MCC2 A0A1A8D6A3 A0A1A8I6Y7 A0A1A8JLT0 A0A1A8PXN1 A0A1A8LDX9 A0A3B3DQ97 H3BBI3 A0A1A8VJG5 A0A1A8NLV4 A0A1A8FDN3 A0A1A8BK93 E9FVE8 A0A0P4XQ40 A0A1A8AE14 A0A1A8S7Y5 A0A3B1JRV5 A0A1A8Q9I1 A0A3B4BMT1 A0A3Q3K2L4 H3D1P9 A0A3B4EAE7 A0A0P6DYW3 H2UZ09 A0A1A8J1I8 A0A3B1JT66 W5U9E7 A0A1A8UMS9 A0A2L2YBS1 A0A0N8ASY2 A0A0P5LCU7 A0A1A8PCB0 A0A1A8KCG1 A0A1A8MU89 A0A1A8CNE5 A0A1A8GLA4 A0A0P4ZGI8 A0A1S3HM79 A0A3B4EV49 A0A3Q2VKF6 A0A3P8Q3U5 A0A3P9BKA9 A0A3Q1IY29
PDB
5BXH
E-value=8.83567e-29,
Score=313
Ontologies
GO
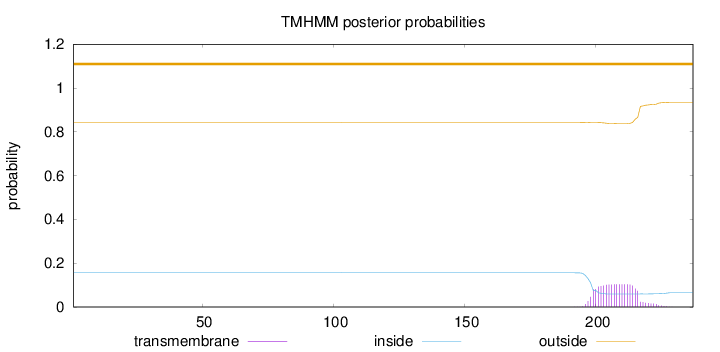
Topology
Length:
237
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.97084
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.15649
outside
1 - 237
Population Genetic Test Statistics
Pi
392.428029
Theta
209.056525
Tajima's D
2.761409
CLR
0.000709
CSRT
0.965451727413629
Interpretation
Uncertain
