Gene
KWMTBOMO15671
Pre Gene Modal
BGIBMGA002292
Annotation
hypothetical_protein_KGM_17319_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.106
Sequence
CDS
ATGGACGTGGGACGCTTAATCGAATGCGTCAAAGACCACGTGTACTTATACGATATAAAACATAAGGATTACAAAAACATCGCACTCAAAACGGCCGTGTGGGAAGCCATCGCGAAGGAATTACATCAAACATGCGATACCGTGAAATTAAAATGGAAAACGATAAGAGATGGGTATATAAAATACAAAAAACAGGTTAAGGATAGCGCCTTCAACGGTAAAAAGATAAACGACTACATATGGAGATCGCAATTAGTTTTTTTGGACAGTCACAACGTGACAAGAAACTCCAAGGCGAATCAGAATAATCAAAAGGCTTTAACGCAGCTACAAACCGAAGATTCGCCCGGTGAGGCAGCTTGGCACTCACCAAAACCCGAAGCGCACACATCCCAAGAATCGCAAAGCCCGTCCGATATGAATTATCAAAATGAATCGAGACTGACCAAGCGCAAGGCTGAAGACGACGTGGACAAAATTCTAAATTATTTAGTTAACAAAAAAGAAAAAAAATATGATGCCGTTGATTATCTGTTTATGAGTTACGCGGAAACGTTCAAGACCTTCTCGTCTCGAACTCAAACCGATATGAAATTGGAATTAGCTAATTTATTTCTTAACGCCGAACTAAAAGAGTTTGCGAACGCCGAAAGCGCTCAGAGACTGACCGCAGAGGCCCAGTCCGGTTCGGATAGTTCCAGTAGCTCAAGCGATTCGGATAACTGCGAGAATTCTCAAACAGATCCCATAGGAACGATGGATGTTAAAATTGAAAGGTCATCGACGCCTGATTACTTTGAGCAGAAATTTTAG
Protein
MDVGRLIECVKDHVYLYDIKHKDYKNIALKTAVWEAIAKELHQTCDTVKLKWKTIRDGYIKYKKQVKDSAFNGKKINDYIWRSQLVFLDSHNVTRNSKANQNNQKALTQLQTEDSPGEAAWHSPKPEAHTSQESQSPSDMNYQNESRLTKRKAEDDVDKILNYLVNKKEKKYDAVDYLFMSYAETFKTFSSRTQTDMKLELANLFLNAELKEFANAESAQRLTAEAQSGSDSSSSSSDSDNCENSQTDPIGTMDVKIERSSTPDYFEQKF
Summary
Uniprot
H9IYF8
A0A212FGZ0
A0A2H1VF98
A0A2A4K3A8
A0A3S2M6U7
J9JRQ0
+ More
A0A2W1BU83 A0A0L7L5B2 A0A1E1WE40 A0A3S2P7I9 A0A3S2NRA7 A0A212FEZ1 X1WYJ6 J9LVY4 A0A3S2NZB4 A0A2M4BZC7 A0A2M4BZ16 E2BIE9 A0A0K8SBQ4 A0A1I8P6E0 A0A2M3ZJ02 A0A2H1W7J3 A0A1I8MLD8 W8BQJ5 A0A2M4AW94 A0A3S2M078 A0A2A4JTL3 A0A2M4BZ88 A0A2M4BYS0 A0A146KXF2 A0A0A9ZCL6 A0A2M4BYY0 A0A2A4JPR3 A0A194R4S6 A0A2W1BNH7 A0A2H1V9R7 A0A2S2QT08 A0A1A8J5L9 A0A1A8RMH5 A0A194QAK9 A0A2S2NV09 A0A182JJQ2 J9KY88 E7F8Q9 A0A194R5M9 A0A1A8BJ41 A0A1A8LKB3
A0A2W1BU83 A0A0L7L5B2 A0A1E1WE40 A0A3S2P7I9 A0A3S2NRA7 A0A212FEZ1 X1WYJ6 J9LVY4 A0A3S2NZB4 A0A2M4BZC7 A0A2M4BZ16 E2BIE9 A0A0K8SBQ4 A0A1I8P6E0 A0A2M3ZJ02 A0A2H1W7J3 A0A1I8MLD8 W8BQJ5 A0A2M4AW94 A0A3S2M078 A0A2A4JTL3 A0A2M4BZ88 A0A2M4BYS0 A0A146KXF2 A0A0A9ZCL6 A0A2M4BYY0 A0A2A4JPR3 A0A194R4S6 A0A2W1BNH7 A0A2H1V9R7 A0A2S2QT08 A0A1A8J5L9 A0A1A8RMH5 A0A194QAK9 A0A2S2NV09 A0A182JJQ2 J9KY88 E7F8Q9 A0A194R5M9 A0A1A8BJ41 A0A1A8LKB3
Pubmed
EMBL
BABH01020485
AGBW02008579
OWR53002.1
ODYU01002266
SOQ39523.1
NWSH01000176
+ More
PCG78741.1 RSAL01000023 RVE52308.1 RVE52311.1 ABLF02041152 ABLF02041159 KZ149935 PZC77194.1 JTDY01002843 KOB70615.1 GDQN01005800 JAT85254.1 RSAL01000006 RVE54331.1 RVE54330.1 AGBW02008891 OWR52290.1 ABLF02013213 ABLF02013216 ABLF02007738 ABLF02007739 RSAL01000084 RVE48389.1 GGFJ01009110 MBW58251.1 GGFJ01009111 MBW58252.1 GL448504 EFN84537.1 GBRD01015322 JAG50504.1 GGFM01007773 MBW28524.1 ODYU01006520 SOQ48444.1 GAMC01003045 JAC03511.1 GGFK01011714 MBW45035.1 RSAL01000090 RVE48089.1 NWSH01000604 PCG75377.1 GGFJ01009070 MBW58211.1 GGFJ01009069 MBW58210.1 GDHC01017486 JAQ01143.1 GBHO01002521 GBHO01002519 GBRD01015978 GDHC01005923 JAG41083.1 JAG41085.1 JAG49848.1 JAQ12706.1 GGFJ01009071 MBW58212.1 NWSH01000885 PCG73698.1 KQ460685 KPJ12818.1 KZ150007 PZC75184.1 ODYU01001420 SOQ37578.1 GGMS01011661 MBY80864.1 HAED01018472 SBR04917.1 HAEH01018199 SBS07320.1 KQ459232 KPJ02469.1 GGMR01007947 MBY20566.1 AXCP01009338 ABLF02017539 CT030204 KPJ12819.1 HADZ01003278 SBP67219.1 HAEF01006957 SBR44339.1
PCG78741.1 RSAL01000023 RVE52308.1 RVE52311.1 ABLF02041152 ABLF02041159 KZ149935 PZC77194.1 JTDY01002843 KOB70615.1 GDQN01005800 JAT85254.1 RSAL01000006 RVE54331.1 RVE54330.1 AGBW02008891 OWR52290.1 ABLF02013213 ABLF02013216 ABLF02007738 ABLF02007739 RSAL01000084 RVE48389.1 GGFJ01009110 MBW58251.1 GGFJ01009111 MBW58252.1 GL448504 EFN84537.1 GBRD01015322 JAG50504.1 GGFM01007773 MBW28524.1 ODYU01006520 SOQ48444.1 GAMC01003045 JAC03511.1 GGFK01011714 MBW45035.1 RSAL01000090 RVE48089.1 NWSH01000604 PCG75377.1 GGFJ01009070 MBW58211.1 GGFJ01009069 MBW58210.1 GDHC01017486 JAQ01143.1 GBHO01002521 GBHO01002519 GBRD01015978 GDHC01005923 JAG41083.1 JAG41085.1 JAG49848.1 JAQ12706.1 GGFJ01009071 MBW58212.1 NWSH01000885 PCG73698.1 KQ460685 KPJ12818.1 KZ150007 PZC75184.1 ODYU01001420 SOQ37578.1 GGMS01011661 MBY80864.1 HAED01018472 SBR04917.1 HAEH01018199 SBS07320.1 KQ459232 KPJ02469.1 GGMR01007947 MBY20566.1 AXCP01009338 ABLF02017539 CT030204 KPJ12819.1 HADZ01003278 SBP67219.1 HAEF01006957 SBR44339.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
CDD
ProteinModelPortal
H9IYF8
A0A212FGZ0
A0A2H1VF98
A0A2A4K3A8
A0A3S2M6U7
J9JRQ0
+ More
A0A2W1BU83 A0A0L7L5B2 A0A1E1WE40 A0A3S2P7I9 A0A3S2NRA7 A0A212FEZ1 X1WYJ6 J9LVY4 A0A3S2NZB4 A0A2M4BZC7 A0A2M4BZ16 E2BIE9 A0A0K8SBQ4 A0A1I8P6E0 A0A2M3ZJ02 A0A2H1W7J3 A0A1I8MLD8 W8BQJ5 A0A2M4AW94 A0A3S2M078 A0A2A4JTL3 A0A2M4BZ88 A0A2M4BYS0 A0A146KXF2 A0A0A9ZCL6 A0A2M4BYY0 A0A2A4JPR3 A0A194R4S6 A0A2W1BNH7 A0A2H1V9R7 A0A2S2QT08 A0A1A8J5L9 A0A1A8RMH5 A0A194QAK9 A0A2S2NV09 A0A182JJQ2 J9KY88 E7F8Q9 A0A194R5M9 A0A1A8BJ41 A0A1A8LKB3
A0A2W1BU83 A0A0L7L5B2 A0A1E1WE40 A0A3S2P7I9 A0A3S2NRA7 A0A212FEZ1 X1WYJ6 J9LVY4 A0A3S2NZB4 A0A2M4BZC7 A0A2M4BZ16 E2BIE9 A0A0K8SBQ4 A0A1I8P6E0 A0A2M3ZJ02 A0A2H1W7J3 A0A1I8MLD8 W8BQJ5 A0A2M4AW94 A0A3S2M078 A0A2A4JTL3 A0A2M4BZ88 A0A2M4BYS0 A0A146KXF2 A0A0A9ZCL6 A0A2M4BYY0 A0A2A4JPR3 A0A194R4S6 A0A2W1BNH7 A0A2H1V9R7 A0A2S2QT08 A0A1A8J5L9 A0A1A8RMH5 A0A194QAK9 A0A2S2NV09 A0A182JJQ2 J9KY88 E7F8Q9 A0A194R5M9 A0A1A8BJ41 A0A1A8LKB3
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
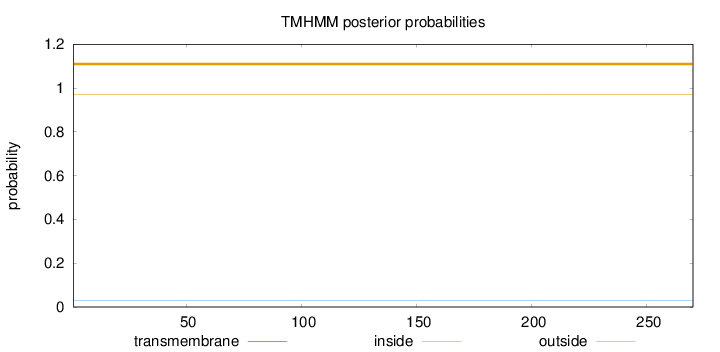
Length:
270
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03103
outside
1 - 270
Population Genetic Test Statistics
Pi
161.4466
Theta
159.422094
Tajima's D
0.453306
CLR
0.034926
CSRT
0.500924953752312
Interpretation
Uncertain
