Gene
KWMTBOMO15668
Pre Gene Modal
BGIBMGA002287
Annotation
PREDICTED:_5-hydroxytryptamine_receptor_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.928
Sequence
CDS
ATGGCGACTTCAAATAATAAATCCCACTCACTGCCTAGCTTTATCGCTTTACCTTCGAGCCTTCTCATACTGGTGTTCATAGTTCTTTTGCAAAACAACGTTGCGGCCACATCTGATTTCACAGAAACATCCCCGGAATACGATTACATATTGACCAATAACTCGACCCTGAACTGGACGTTTTTCGAAGGAAACGCCAGTCACAAGCCGGTGCACATAAAACATTCAAAATACTCCGCCCCCGTTTCAATTTTCTTATCATTGTTGTTTTTGGTCGTGATAGTTGGAACGATTGTTGGTAATATCCTAGTTTGTGTGGCCGTTCGACTGGTCAGAAAATTGAGGAGGCCGAGTAATTATTTAATAGTATCACTGGCGGTCAGTGATCTGTGTGTCGCAATCATAGTCATGCCAGTAGCCACTGTATACGATATAATGGGATCTTGGCCCTTTGGACCCGTAATTTGCGACTTTTGGGTATCGAGTGATGTTTTGTCGTGCACCGCAAGCATCCTGAACCTCTGCATGATCTCAGTGGATCGATACTACGCGATTACAAAACCATTAGAATACGGCGTGAAGAGGACACCGAGGAGGATGCTATTTTGTGTTTTCATAGTATGGATAAGTGCAGCTTTCATTTCTTTACCACCAGTTTTAATACTAGGCAACGAGAAGACTGAGACCTCGTGTTCTGTTTCCCAAAATCAGTATTACCAAGTTTATGCTACGTTAGGATCCTTCTATATCCCGTTGACTGTTATGGTAATTGTGTATTACAAAATATTCAGAGCGGCAAGAAAAATAGTGAAAGATGAGAAACGTGCTCAATCGCATTTAGAAGCGCATTGCTATCTCGAGATTAACGTTAAGAATGGAGGAGCGACCGAAGCGAAACTTCTCGGCGCCCAGACCACGACTCAACCGCCAAGAGGATCTACAGCGAGCACTAACACAACTTGCAGCGTTGAGAAAACCGAAAGCGGGATTGGACGGTGCTTTAGTGGCCAGCGGAAATCAAATGAATCCCAGTACCCGATGCTTCACCAACCAAAGACCCCAACTAAACCTATTCACACTATAAATCGATCTCCATCTACACTAACACCAAACAACAAGCTATTAGTTAAAGAACGAAGTCGCGAGTCGTCGGAGAGTAGTAACAAGATAAATACAAACAGAATTCGCTCATCCCTCTCTAATTTTGCTCAAAAAAGTCATAAAGCCAAAGATATGTTCCAAACGCAGTCGCCGCATCAAAAGAAGCTGCGGTTTCAACTGGCGAAAGAGCGGAAAGCCTCCACGACCCTCGGGATTATTATGTCAGCGTTTATAGTTTGCTGGTTACCGTTTTTCGTGCTGGCCTTACTCAGGCCGTTCCTGAACGAGGACGCTATCCCGGATGCAGTGAGCGCTCTATTCCTCTGGCTGGGCTATTTAAACAGCCTACTCAATCCAATCATTTACGCGACATTAAACAGGGACTTTCGGAAGCCTTTCCAAGAGATTCTGTTCTTTCGATGCTCGAACCTGAACCACATGATGCGCGAGGAGTTCTACCACAGCCAGTACGGAGATCCGGACCATTACTGTGTAAACAACACGACGAAGCTAAATAACTACGACGAAGGAGTCGAAATAGTTTCGGCTGTCGACAGAGAGGAGACAAGAGCATCAGAGAGCTTTCTATGA
Protein
MATSNNKSHSLPSFIALPSSLLILVFIVLLQNNVAATSDFTETSPEYDYILTNNSTLNWTFFEGNASHKPVHIKHSKYSAPVSIFLSLLFLVVIVGTIVGNILVCVAVRLVRKLRRPSNYLIVSLAVSDLCVAIIVMPVATVYDIMGSWPFGPVICDFWVSSDVLSCTASILNLCMISVDRYYAITKPLEYGVKRTPRRMLFCVFIVWISAAFISLPPVLILGNEKTETSCSVSQNQYYQVYATLGSFYIPLTVMVIVYYKIFRAARKIVKDEKRAQSHLEAHCYLEINVKNGGATEAKLLGAQTTTQPPRGSTASTNTTCSVEKTESGIGRCFSGQRKSNESQYPMLHQPKTPTKPIHTINRSPSTLTPNNKLLVKERSRESSESSNKINTNRIRSSLSNFAQKSHKAKDMFQTQSPHQKKLRFQLAKERKASTTLGIIMSAFIVCWLPFFVLALLRPFLNEDAIPDAVSALFLWLGYLNSLLNPIIYATLNRDFRKPFQEILFFRCSNLNHMMREEFYHSQYGDPDHYCVNNTTKLNNYDEGVEIVSAVDREETRASESFL
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
S4UQW2
A0A2W1BJ90
A0A3G6ITM7
A0A2H1VLX3
A0A1E1WD65
A0A0L7KL42
+ More
A0A0N1IGJ9 A0A142F420 A0A194QIH1 D6X0T4 N1NSY0 A0A0C9Q5G1 A0A1Z2RSH1 Q0P7L6 A0A1B6DW61 J3JXM6 K7IM91 A0A3L8E1P0 A0A2H8TQD5 U5XHJ3 B4NH14 A0A1B3P5V7 A0A0P6EWS5 A0A0P5RJL6 A0A0P5N9N9 A0A0N8E489 A0A0P5QUK3 A0A0P5J6M4 A0A0P5XR33 A0A0P4XI68 A0A0P5M379 A0A0P5UF56 A0A0P5WAF6 A0A0P6F8E8 A0A0P5C2K3 A0A0N8D387 A0A0P5TKN2 A0A2S2R0C3 J9M088 A0A0P6FA72 A0A0P5JGC0 A0A0P5PR83 A0A0P5HRW1 A0A0N8C2N5 A0A0P5WEW4 A0A0P5PVQ0 A0A0P5ZH32 A0A0P6F4I8 A0A0N8CYU5 T1FH73 A0A3S4QB33 A0A0P5A957
A0A0N1IGJ9 A0A142F420 A0A194QIH1 D6X0T4 N1NSY0 A0A0C9Q5G1 A0A1Z2RSH1 Q0P7L6 A0A1B6DW61 J3JXM6 K7IM91 A0A3L8E1P0 A0A2H8TQD5 U5XHJ3 B4NH14 A0A1B3P5V7 A0A0P6EWS5 A0A0P5RJL6 A0A0P5N9N9 A0A0N8E489 A0A0P5QUK3 A0A0P5J6M4 A0A0P5XR33 A0A0P4XI68 A0A0P5M379 A0A0P5UF56 A0A0P5WAF6 A0A0P6F8E8 A0A0P5C2K3 A0A0N8D387 A0A0P5TKN2 A0A2S2R0C3 J9M088 A0A0P6FA72 A0A0P5JGC0 A0A0P5PR83 A0A0P5HRW1 A0A0N8C2N5 A0A0P5WEW4 A0A0P5PVQ0 A0A0P5ZH32 A0A0P6F4I8 A0A0N8CYU5 T1FH73 A0A3S4QB33 A0A0P5A957
Pubmed
EMBL
JX878498
AGL46976.1
KZ150086
PZC73775.1
MH025798
AZA07970.1
+ More
ODYU01003281 SOQ41835.1 GDQN01006126 JAT84928.1 JTDY01009120 KOB64083.1 KQ459941 KPJ19176.1 KT946790 AMQ67549.1 KQ458761 KPJ05194.1 KQ971372 EFA10682.2 BK005867 DAA64504.1 GBYB01009273 JAG79040.1 KX360127 ASA47150.1 AM076717 CAJ28210.1 GEDC01007397 JAS29901.1 BT127996 AEE62958.1 QOIP01000001 RLU26305.1 GFXV01003663 MBW15468.1 KF297572 AGZ62050.1 CH964272 EDW84511.1 KU712068 AOG12998.1 GDIQ01056998 GDIQ01056997 JAN37740.1 GDIQ01203954 GDIQ01101592 JAL50134.1 GDIQ01148924 GDIQ01049859 JAL02802.1 GDIQ01172159 GDIQ01172158 GDIQ01130986 GDIQ01128433 GDIQ01125690 GDIQ01120780 GDIQ01063356 GDIQ01059600 GDIQ01059599 GDIQ01044905 GDIQ01032820 JAN31381.1 GDIQ01229247 GDIQ01132340 GDIQ01129680 GDIQ01063355 GDIQ01048366 JAL22046.1 GDIQ01203953 GDIP01044650 GDIQ01075495 GDIQ01072645 GDIQ01072644 GDIQ01051263 JAK47772.1 JAM59065.1 GDIP01070593 JAM33122.1 GDIP01240830 JAI82571.1 GDIP01196888 GDIP01171125 GDIQ01161417 GDIQ01129679 GDIP01123941 GDIP01088890 GDIQ01044904 JAJ26514.1 JAK90308.1 GDIP01113949 JAL89765.1 GDIP01179077 GDIP01112692 GDIP01109885 GDIP01095543 GDIP01088891 JAM14824.1 GDIQ01051262 JAN43475.1 GDIP01198429 GDIP01176445 GDIP01136834 GDIP01130587 GDIP01073152 JAJ46957.1 GDIP01073151 JAM30564.1 GDIP01124994 JAL78720.1 GGMS01014037 MBY83240.1 ABLF02033750 ABLF02033754 ABLF02033757 ABLF02033759 ABLF02033760 GDIQ01063354 JAN31383.1 GDIQ01199804 JAK51921.1 GDIQ01130985 JAL20741.1 GDIQ01223830 GDIQ01220931 JAK27895.1 GDIQ01231492 GDIQ01229246 GDIQ01132339 GDIQ01120779 GDIQ01063353 JAL30947.1 GDIP01176446 GDIP01130588 GDIP01112693 GDIP01087792 GDIP01061148 JAM15923.1 GDIQ01129678 JAL22048.1 GDIP01044651 JAM59064.1 GDIQ01061658 JAN33079.1 GDIP01085487 JAM18228.1 AMQM01007726 KB097661 ESN92183.1 NCKU01009845 RWS01106.1 GDIP01202376 JAJ21026.1
ODYU01003281 SOQ41835.1 GDQN01006126 JAT84928.1 JTDY01009120 KOB64083.1 KQ459941 KPJ19176.1 KT946790 AMQ67549.1 KQ458761 KPJ05194.1 KQ971372 EFA10682.2 BK005867 DAA64504.1 GBYB01009273 JAG79040.1 KX360127 ASA47150.1 AM076717 CAJ28210.1 GEDC01007397 JAS29901.1 BT127996 AEE62958.1 QOIP01000001 RLU26305.1 GFXV01003663 MBW15468.1 KF297572 AGZ62050.1 CH964272 EDW84511.1 KU712068 AOG12998.1 GDIQ01056998 GDIQ01056997 JAN37740.1 GDIQ01203954 GDIQ01101592 JAL50134.1 GDIQ01148924 GDIQ01049859 JAL02802.1 GDIQ01172159 GDIQ01172158 GDIQ01130986 GDIQ01128433 GDIQ01125690 GDIQ01120780 GDIQ01063356 GDIQ01059600 GDIQ01059599 GDIQ01044905 GDIQ01032820 JAN31381.1 GDIQ01229247 GDIQ01132340 GDIQ01129680 GDIQ01063355 GDIQ01048366 JAL22046.1 GDIQ01203953 GDIP01044650 GDIQ01075495 GDIQ01072645 GDIQ01072644 GDIQ01051263 JAK47772.1 JAM59065.1 GDIP01070593 JAM33122.1 GDIP01240830 JAI82571.1 GDIP01196888 GDIP01171125 GDIQ01161417 GDIQ01129679 GDIP01123941 GDIP01088890 GDIQ01044904 JAJ26514.1 JAK90308.1 GDIP01113949 JAL89765.1 GDIP01179077 GDIP01112692 GDIP01109885 GDIP01095543 GDIP01088891 JAM14824.1 GDIQ01051262 JAN43475.1 GDIP01198429 GDIP01176445 GDIP01136834 GDIP01130587 GDIP01073152 JAJ46957.1 GDIP01073151 JAM30564.1 GDIP01124994 JAL78720.1 GGMS01014037 MBY83240.1 ABLF02033750 ABLF02033754 ABLF02033757 ABLF02033759 ABLF02033760 GDIQ01063354 JAN31383.1 GDIQ01199804 JAK51921.1 GDIQ01130985 JAL20741.1 GDIQ01223830 GDIQ01220931 JAK27895.1 GDIQ01231492 GDIQ01229246 GDIQ01132339 GDIQ01120779 GDIQ01063353 JAL30947.1 GDIP01176446 GDIP01130588 GDIP01112693 GDIP01087792 GDIP01061148 JAM15923.1 GDIQ01129678 JAL22048.1 GDIP01044651 JAM59064.1 GDIQ01061658 JAN33079.1 GDIP01085487 JAM18228.1 AMQM01007726 KB097661 ESN92183.1 NCKU01009845 RWS01106.1 GDIP01202376 JAJ21026.1
Proteomes
Pfam
PF00001 7tm_1
Interpro
ProteinModelPortal
S4UQW2
A0A2W1BJ90
A0A3G6ITM7
A0A2H1VLX3
A0A1E1WD65
A0A0L7KL42
+ More
A0A0N1IGJ9 A0A142F420 A0A194QIH1 D6X0T4 N1NSY0 A0A0C9Q5G1 A0A1Z2RSH1 Q0P7L6 A0A1B6DW61 J3JXM6 K7IM91 A0A3L8E1P0 A0A2H8TQD5 U5XHJ3 B4NH14 A0A1B3P5V7 A0A0P6EWS5 A0A0P5RJL6 A0A0P5N9N9 A0A0N8E489 A0A0P5QUK3 A0A0P5J6M4 A0A0P5XR33 A0A0P4XI68 A0A0P5M379 A0A0P5UF56 A0A0P5WAF6 A0A0P6F8E8 A0A0P5C2K3 A0A0N8D387 A0A0P5TKN2 A0A2S2R0C3 J9M088 A0A0P6FA72 A0A0P5JGC0 A0A0P5PR83 A0A0P5HRW1 A0A0N8C2N5 A0A0P5WEW4 A0A0P5PVQ0 A0A0P5ZH32 A0A0P6F4I8 A0A0N8CYU5 T1FH73 A0A3S4QB33 A0A0P5A957
A0A0N1IGJ9 A0A142F420 A0A194QIH1 D6X0T4 N1NSY0 A0A0C9Q5G1 A0A1Z2RSH1 Q0P7L6 A0A1B6DW61 J3JXM6 K7IM91 A0A3L8E1P0 A0A2H8TQD5 U5XHJ3 B4NH14 A0A1B3P5V7 A0A0P6EWS5 A0A0P5RJL6 A0A0P5N9N9 A0A0N8E489 A0A0P5QUK3 A0A0P5J6M4 A0A0P5XR33 A0A0P4XI68 A0A0P5M379 A0A0P5UF56 A0A0P5WAF6 A0A0P6F8E8 A0A0P5C2K3 A0A0N8D387 A0A0P5TKN2 A0A2S2R0C3 J9M088 A0A0P6FA72 A0A0P5JGC0 A0A0P5PR83 A0A0P5HRW1 A0A0N8C2N5 A0A0P5WEW4 A0A0P5PVQ0 A0A0P5ZH32 A0A0P6F4I8 A0A0N8CYU5 T1FH73 A0A3S4QB33 A0A0P5A957
PDB
6CM4
E-value=5.07664e-48,
Score=484
Ontologies
GO
Topology
Subcellular location
Cell membrane
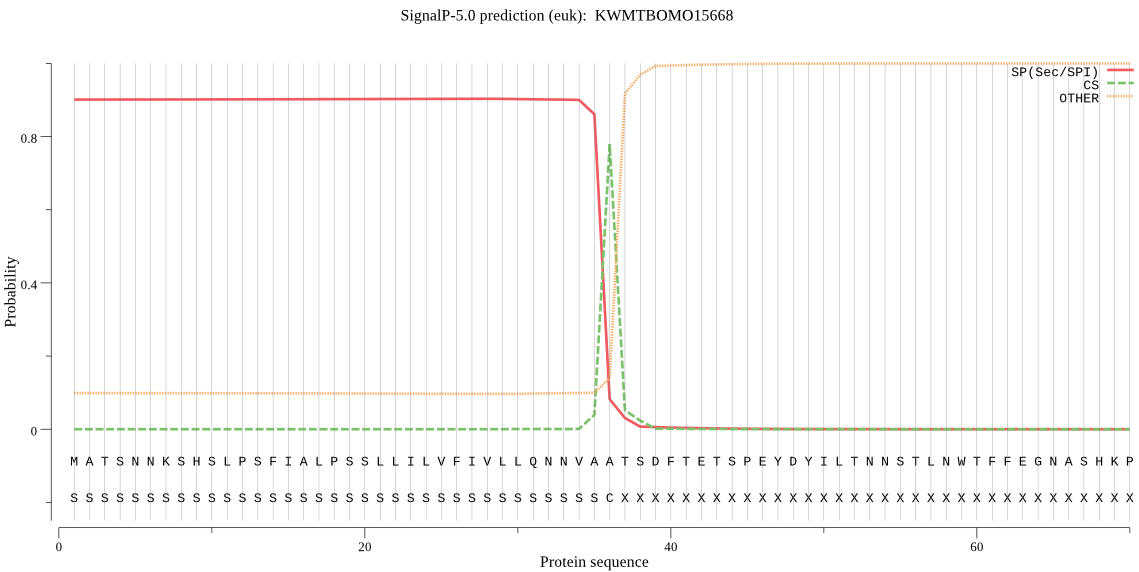
SignalP
Position: 1 - 36,
Likelihood: 0.903184
Length:
563
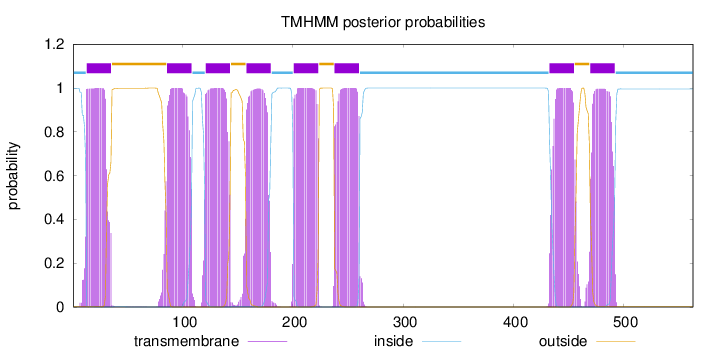
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
178.39255
Exp number, first 60 AAs:
20.90068
Total prob of N-in:
0.99862
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 85
TMhelix
86 - 108
inside
109 - 120
TMhelix
121 - 143
outside
144 - 157
TMhelix
158 - 180
inside
181 - 200
TMhelix
201 - 223
outside
224 - 237
TMhelix
238 - 260
inside
261 - 432
TMhelix
433 - 455
outside
456 - 469
TMhelix
470 - 492
inside
493 - 563
Population Genetic Test Statistics
Pi
39.771251
Theta
34.854331
Tajima's D
0.358721
CLR
0.923671
CSRT
0.482625868706565
Interpretation
Uncertain
