Gene
KWMTBOMO15664 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002288
Annotation
C-type_lectin_21_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.647 Extracellular Reliability : 1.151
Sequence
CDS
ATGCGGGATATCAACGGCTGGTTGAAGCTTCAAGAGATCCCAGCTACCTGGCAGGAGGCTCGATTGAGATGTCATCTTGAAGGGTCCGTTTTGGCATCTCCTCTGGACGATGCGTTGAAGAGCGGCATGCTATCTCTAATAAAGAACAAGAAGACTTCATGTGGTATATTCACCGGCATCCATGCGACATTTTCTAAAGGAGACTACCGTTCTGTTGAAGGTGTCCCATTGGCGAAAATTCCTCACGATTGGGCGGATTATGAACCAGATAATGCTGGAGGTGACGAAAACTGTATCCTCATGAACCCTGATGGAAACTTCGCCGATGTTAATTGCACTGAAACGTTCCAATACGTTTGCTATAAGAAAAAGACTTCGACTATTGCTATGGCTTCTTGTGGCAGTGTTGATAGCGAATATGTTCTCAGCAAAGACACTGGTAATTGTTACAAGTTCCACAAAGTTCCCCGTACCTGGTCGCGTGCCTACATGGCCTGTTCAGCTGAAGGCGGATACCTCACGATTATTAACAACGAGAAGGAGGCCGTGTTCCTAAGGGACCTTTTCGTGAAGAACCCTGCCGTTCACATGATCGGAAGTTTCTGGAAAGACGTTGCCTTCATTGGCTTCCACGACTGGAATGAGCGCGGGGAATGGTTAACGATCAATGGTGAGAAATTACAAGAGGCCGGTTACGAGAAATGGTCTGGCGGCGAGCCTAGTAACGCGACGACTGGTGAATATTGCGGCTCTATCTACCGCTCGGCTCTACTCAACGACCTGTGGTGCGAAAAGCCGGCACCGTTTATCTGCGAGAAGGAACCTCGCAGCTTACTCCGAGAGCACGACGACAAATGA
Protein
MRDINGWLKLQEIPATWQEARLRCHLEGSVLASPLDDALKSGMLSLIKNKKTSCGIFTGIHATFSKGDYRSVEGVPLAKIPHDWADYEPDNAGGDENCILMNPDGNFADVNCTETFQYVCYKKKTSTIAMASCGSVDSEYVLSKDTGNCYKFHKVPRTWSRAYMACSAEGGYLTIINNEKEAVFLRDLFVKNPAVHMIGSFWKDVAFIGFHDWNERGEWLTINGEKLQEAGYEKWSGGEPSNATTGEYCGSIYRSALLNDLWCEKPAPFICEKEPRSLLREHDDK
Summary
Uniprot
O97363
H9IYF4
D2X2F6
H9IYF5
A0A194QAQ3
A0A0N1IPT5
+ More
A0A3S2P588 A0A096ZGV0 Q9NBV9 G5CJE6 C0L7M0 C0L7M1 A0A3S2NP58 A0A2W1BVD9 A0A2A4IYF0 A0A2A4J280 A0A2H1WK53 Q19AB1 A0A2W1BYL2 I1VR05 A0A2W1BZP3 A0A212FF49 A0A173M1W5 A0A1E1WP12 A0A096ZGV4 A0A3S2L865 A0A0N1PJ18 A0A2A4J242 A0A2A4J3T7 A0A2W1BVG4 A0A1E1WQ86 A0A2W1BTF0 A0A2W1C0J5 B7SVM1 A0A0L7LUC3 R9UPV0 A0A0D3M5W6 O96359 A0A194QAQ8 A0A2Z5WQ31 Q5MGF0 A0A3S2TGZ3 A0A2A4IWU7 A0A0L7KKI4 A0A0L7KUN5 A0A1E1W152 A0A3S2NEY7 A0A2W1BTE2 A0A2A4J4S6 Q5UAW8 A0A0L7LBS8 Q5UAW7 A0A219YXH0 A0A2A4J2P8 A0A219YXH1 A0A3S2LAT9 A0A0L7KS66 A0A219YXK5 Q5MGG4 A0A219YXH2 B1A5Z5 A0A3G1HH34 A0A3S2NK24 A0A219YXG9 A0A2W1BF95 I1VR07 A0A219YXH4 A0A1C9EGM6 A0A219YXI5 A0A0L7LT05 A0A219YXH3 A0A096ZGV2 A0A2A4K927 A0A219YXI1 A0A2W1BDY0 A0A219YXG7 A0A219YXG8 A0A219YXH5 A0A2H1VQU1 A0A3G1HH41 A0A3G1HH66 A0A219YXI4 A0A219YXI8 A0A3S2TBA6 A0A194QMK1 A0A219YXH8 A0A3G1HH42 A0A219YXH6 I1VR06 I4DJJ2 A0A2W1B2K8 A0A219YXL1 A0A194PEZ5 S4PJZ4
A0A3S2P588 A0A096ZGV0 Q9NBV9 G5CJE6 C0L7M0 C0L7M1 A0A3S2NP58 A0A2W1BVD9 A0A2A4IYF0 A0A2A4J280 A0A2H1WK53 Q19AB1 A0A2W1BYL2 I1VR05 A0A2W1BZP3 A0A212FF49 A0A173M1W5 A0A1E1WP12 A0A096ZGV4 A0A3S2L865 A0A0N1PJ18 A0A2A4J242 A0A2A4J3T7 A0A2W1BVG4 A0A1E1WQ86 A0A2W1BTF0 A0A2W1C0J5 B7SVM1 A0A0L7LUC3 R9UPV0 A0A0D3M5W6 O96359 A0A194QAQ8 A0A2Z5WQ31 Q5MGF0 A0A3S2TGZ3 A0A2A4IWU7 A0A0L7KKI4 A0A0L7KUN5 A0A1E1W152 A0A3S2NEY7 A0A2W1BTE2 A0A2A4J4S6 Q5UAW8 A0A0L7LBS8 Q5UAW7 A0A219YXH0 A0A2A4J2P8 A0A219YXH1 A0A3S2LAT9 A0A0L7KS66 A0A219YXK5 Q5MGG4 A0A219YXH2 B1A5Z5 A0A3G1HH34 A0A3S2NK24 A0A219YXG9 A0A2W1BF95 I1VR07 A0A219YXH4 A0A1C9EGM6 A0A219YXI5 A0A0L7LT05 A0A219YXH3 A0A096ZGV2 A0A2A4K927 A0A219YXI1 A0A2W1BDY0 A0A219YXG7 A0A219YXG8 A0A219YXH5 A0A2H1VQU1 A0A3G1HH41 A0A3G1HH66 A0A219YXI4 A0A219YXI8 A0A3S2TBA6 A0A194QMK1 A0A219YXH8 A0A3G1HH42 A0A219YXH6 I1VR06 I4DJJ2 A0A2W1B2K8 A0A219YXL1 A0A194PEZ5 S4PJZ4
Pubmed
EMBL
AJ011573
CAB38429.1
BABH01020449
GQ487489
ADB12587.1
BABH01020450
+ More
KQ459232 KPJ02552.1 KQ460044 KPJ18317.1 RSAL01000019 RVE52735.1 KF752419 AIR95998.1 AF242202 AAF91316.2 JN133501 AEO52696.1 FJ666210 ACN41858.1 FJ666211 ACN41859.1 RVE52734.1 KZ149926 PZC77594.1 NWSH01005098 PCG64438.1 NWSH01003559 PCG66111.1 ODYU01009180 SOQ53429.1 DQ533877 ABF83203.1 KZ149905 PZC78327.1 JQ269656 AFI47451.1 PZC78330.1 AGBW02008849 OWR52366.1 LC150882 BAV01248.1 GDQN01002397 JAT88657.1 KF752421 AIR96000.1 RSAL01000824 RVE41129.1 KPJ18324.1 NWSH01003594 PCG66071.1 PCG66072.1 PZC78331.1 GDQN01002063 JAT88991.1 PZC78329.1 PZC77593.1 EU770390 ACI32834.1 JTDY01000107 KOB78821.1 KC733761 AGN70857.1 KF285999 AIC33761.1 AF023275 AAD09286.1 KPJ02557.1 LC209794 BBC20960.1 AY829836 AAV91450.1 RSAL01000150 RVE45770.1 PCG64437.1 JTDY01009247 KOB63853.1 JTDY01005614 KOB66830.1 GDQN01010324 JAT80730.1 RVE45766.1 PZC78328.1 NWSH01002983 PCG67147.1 AY768811 AAV41236.1 JTDY01001800 KOB72850.1 AY768812 AAV41237.2 KX550127 AQY54444.1 NWSH01003419 PCG66335.1 KX550126 AQY54443.1 RSAL01000062 RVE49558.1 JTDY01006582 KOB65869.1 KX550129 AQY54446.1 AY829822 AAV91436.1 KX550125 AQY54442.1 EU426550 ABZ81710.1 KY111304 KY111307 AQX37240.1 AQX37243.1 RVE49559.1 KX550120 AQY54437.1 KZ150306 PZC71506.1 JQ269658 AFI47453.1 KX550130 AQY54447.1 KX778610 AON96650.1 KX550141 AQY54458.1 JTDY01000152 KOB78577.1 KX550128 AQY54445.1 KF752420 AIR95999.1 NWSH01000031 PCG80506.1 KX550136 AQY54453.1 PZC71507.1 KX550123 AQY54440.1 KX550122 AQY54439.1 KX550131 AQY54448.1 ODYU01003868 SOQ43168.1 KY111311 AQX37247.1 KY111312 AQX37248.1 KX550138 AQY54455.1 KX550142 AQY54459.1 RSAL01001098 RVE40947.1 KQ461198 KPJ06185.1 KX550124 AQY54441.1 KY111310 AQX37246.1 KX550132 AQY54449.1 JQ269657 AFI47452.1 AK401460 BAM18082.1 KZ150418 PZC70939.1 KX550139 AQY54456.1 KQ459606 KPI91593.1 GAIX01004755 JAA87805.1
KQ459232 KPJ02552.1 KQ460044 KPJ18317.1 RSAL01000019 RVE52735.1 KF752419 AIR95998.1 AF242202 AAF91316.2 JN133501 AEO52696.1 FJ666210 ACN41858.1 FJ666211 ACN41859.1 RVE52734.1 KZ149926 PZC77594.1 NWSH01005098 PCG64438.1 NWSH01003559 PCG66111.1 ODYU01009180 SOQ53429.1 DQ533877 ABF83203.1 KZ149905 PZC78327.1 JQ269656 AFI47451.1 PZC78330.1 AGBW02008849 OWR52366.1 LC150882 BAV01248.1 GDQN01002397 JAT88657.1 KF752421 AIR96000.1 RSAL01000824 RVE41129.1 KPJ18324.1 NWSH01003594 PCG66071.1 PCG66072.1 PZC78331.1 GDQN01002063 JAT88991.1 PZC78329.1 PZC77593.1 EU770390 ACI32834.1 JTDY01000107 KOB78821.1 KC733761 AGN70857.1 KF285999 AIC33761.1 AF023275 AAD09286.1 KPJ02557.1 LC209794 BBC20960.1 AY829836 AAV91450.1 RSAL01000150 RVE45770.1 PCG64437.1 JTDY01009247 KOB63853.1 JTDY01005614 KOB66830.1 GDQN01010324 JAT80730.1 RVE45766.1 PZC78328.1 NWSH01002983 PCG67147.1 AY768811 AAV41236.1 JTDY01001800 KOB72850.1 AY768812 AAV41237.2 KX550127 AQY54444.1 NWSH01003419 PCG66335.1 KX550126 AQY54443.1 RSAL01000062 RVE49558.1 JTDY01006582 KOB65869.1 KX550129 AQY54446.1 AY829822 AAV91436.1 KX550125 AQY54442.1 EU426550 ABZ81710.1 KY111304 KY111307 AQX37240.1 AQX37243.1 RVE49559.1 KX550120 AQY54437.1 KZ150306 PZC71506.1 JQ269658 AFI47453.1 KX550130 AQY54447.1 KX778610 AON96650.1 KX550141 AQY54458.1 JTDY01000152 KOB78577.1 KX550128 AQY54445.1 KF752420 AIR95999.1 NWSH01000031 PCG80506.1 KX550136 AQY54453.1 PZC71507.1 KX550123 AQY54440.1 KX550122 AQY54439.1 KX550131 AQY54448.1 ODYU01003868 SOQ43168.1 KY111311 AQX37247.1 KY111312 AQX37248.1 KX550138 AQY54455.1 KX550142 AQY54459.1 RSAL01001098 RVE40947.1 KQ461198 KPJ06185.1 KX550124 AQY54441.1 KY111310 AQX37246.1 KX550132 AQY54449.1 JQ269657 AFI47452.1 AK401460 BAM18082.1 KZ150418 PZC70939.1 KX550139 AQY54456.1 KQ459606 KPI91593.1 GAIX01004755 JAA87805.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
O97363
H9IYF4
D2X2F6
H9IYF5
A0A194QAQ3
A0A0N1IPT5
+ More
A0A3S2P588 A0A096ZGV0 Q9NBV9 G5CJE6 C0L7M0 C0L7M1 A0A3S2NP58 A0A2W1BVD9 A0A2A4IYF0 A0A2A4J280 A0A2H1WK53 Q19AB1 A0A2W1BYL2 I1VR05 A0A2W1BZP3 A0A212FF49 A0A173M1W5 A0A1E1WP12 A0A096ZGV4 A0A3S2L865 A0A0N1PJ18 A0A2A4J242 A0A2A4J3T7 A0A2W1BVG4 A0A1E1WQ86 A0A2W1BTF0 A0A2W1C0J5 B7SVM1 A0A0L7LUC3 R9UPV0 A0A0D3M5W6 O96359 A0A194QAQ8 A0A2Z5WQ31 Q5MGF0 A0A3S2TGZ3 A0A2A4IWU7 A0A0L7KKI4 A0A0L7KUN5 A0A1E1W152 A0A3S2NEY7 A0A2W1BTE2 A0A2A4J4S6 Q5UAW8 A0A0L7LBS8 Q5UAW7 A0A219YXH0 A0A2A4J2P8 A0A219YXH1 A0A3S2LAT9 A0A0L7KS66 A0A219YXK5 Q5MGG4 A0A219YXH2 B1A5Z5 A0A3G1HH34 A0A3S2NK24 A0A219YXG9 A0A2W1BF95 I1VR07 A0A219YXH4 A0A1C9EGM6 A0A219YXI5 A0A0L7LT05 A0A219YXH3 A0A096ZGV2 A0A2A4K927 A0A219YXI1 A0A2W1BDY0 A0A219YXG7 A0A219YXG8 A0A219YXH5 A0A2H1VQU1 A0A3G1HH41 A0A3G1HH66 A0A219YXI4 A0A219YXI8 A0A3S2TBA6 A0A194QMK1 A0A219YXH8 A0A3G1HH42 A0A219YXH6 I1VR06 I4DJJ2 A0A2W1B2K8 A0A219YXL1 A0A194PEZ5 S4PJZ4
A0A3S2P588 A0A096ZGV0 Q9NBV9 G5CJE6 C0L7M0 C0L7M1 A0A3S2NP58 A0A2W1BVD9 A0A2A4IYF0 A0A2A4J280 A0A2H1WK53 Q19AB1 A0A2W1BYL2 I1VR05 A0A2W1BZP3 A0A212FF49 A0A173M1W5 A0A1E1WP12 A0A096ZGV4 A0A3S2L865 A0A0N1PJ18 A0A2A4J242 A0A2A4J3T7 A0A2W1BVG4 A0A1E1WQ86 A0A2W1BTF0 A0A2W1C0J5 B7SVM1 A0A0L7LUC3 R9UPV0 A0A0D3M5W6 O96359 A0A194QAQ8 A0A2Z5WQ31 Q5MGF0 A0A3S2TGZ3 A0A2A4IWU7 A0A0L7KKI4 A0A0L7KUN5 A0A1E1W152 A0A3S2NEY7 A0A2W1BTE2 A0A2A4J4S6 Q5UAW8 A0A0L7LBS8 Q5UAW7 A0A219YXH0 A0A2A4J2P8 A0A219YXH1 A0A3S2LAT9 A0A0L7KS66 A0A219YXK5 Q5MGG4 A0A219YXH2 B1A5Z5 A0A3G1HH34 A0A3S2NK24 A0A219YXG9 A0A2W1BF95 I1VR07 A0A219YXH4 A0A1C9EGM6 A0A219YXI5 A0A0L7LT05 A0A219YXH3 A0A096ZGV2 A0A2A4K927 A0A219YXI1 A0A2W1BDY0 A0A219YXG7 A0A219YXG8 A0A219YXH5 A0A2H1VQU1 A0A3G1HH41 A0A3G1HH66 A0A219YXI4 A0A219YXI8 A0A3S2TBA6 A0A194QMK1 A0A219YXH8 A0A3G1HH42 A0A219YXH6 I1VR06 I4DJJ2 A0A2W1B2K8 A0A219YXL1 A0A194PEZ5 S4PJZ4
PDB
5G6U
E-value=3.76971e-08,
Score=137
Ontologies
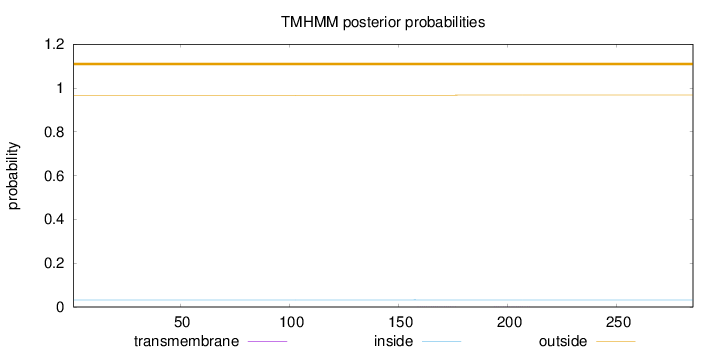
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01046
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.03237
outside
1 - 285
Population Genetic Test Statistics
Pi
224.692914
Theta
195.891302
Tajima's D
0.354184
CLR
0.014686
CSRT
0.47637618119094
Interpretation
Uncertain
