Gene
KWMTBOMO15659
Pre Gene Modal
BGIBMGA000827
Annotation
PREDICTED:_glycerate_kinase_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.197 Mitochondrial Reliability : 1.497
Sequence
CDS
ATGTCCAGAGAAGTAGAAAATATTTTACAATCTAAAATTAAATATGGTATCATAAGCATTCCTATGGGAAGTTTGGACGTATTTAACAAATCAAGAAACGTTGAGTATTTTGAAGGAGCTAAAGATAATTTACCTGACAATAGTGCACAAAACACTGCATTGAAAATAAAGAATCTCATTACACAATTAAACAAAGATGATCTATTACTAGTACTTATTTCAGGAGGTGGTTCAGCATTACTGCCTTTGCCAAAATCACCAATAACTTTAGAAGAAAAAATTGGACTTGTAAAGAAGTTGGCTAATTCTGGTGCTGATATTAAAGAATTAAATACTGTTAGGAAAGTAATATCAGATTTAAAAGGAGGCCAACTGGCAGTAAAGGCACAACCTGCTCAAGTTGTCTCACTAATCCTTTCTGATATTGTTGGTGACCCTTTAGATCTTATAGCAAGTGGTCCTACTGTACAGAATACTGATGGAGCTAATAAAGCAGTTGATGTTTTAAAAAAATACAATTTAATAGATGCACTACCTAAATCAGTGCAAACATTACTTGAAAATAATGGAGATAATTTAGTATTTCCAACAAATAATACATCAAATTACATAATTGGTTCAAACAAAATAAGCACTAAAGCTGCAGTTGTACAGTGTATAGAACTTAATTACCTACCATTAGTACTATCAAATAAAGTTACTGGAAATGTACAAGATGTAGCAAACAAATATAGTAAACTAGTGACCGTAATTTGTAAATATTTGAGGCAAAATTTGGAAATTGATGAATTAAAATCAAACATAAAAAAACTGGAAATATCTGGAACAGATTTAAAAGTTTTGAATGAAATAAAAATAAGTAACAAGAAACCGTTATGTCTTATACTTGGAGGAGAGATAACTGTAGCAGTAAAAGGAACGGGTAAGGGTGGTAGAAATCAACAACTGGCTTTGGAATTTTCTAAATATCTACACAAAGTCAAAGATCAATTGAATGACTTTGATATATTTATACTGAGTGCTGGAACTGATGGAATAGATGGCCCAACTGATGCAGCAGGGGCGATAGGATATTTGAATCTCATTTCAGAATCTACAGCTGATGGTTTAGATGTGGATAAATATTTGGCCAATAATGATACCTACAATTTCTTTAAATTATTTAAAAATGACCATTTACATGTATTTACTGGTCACACCAATACTAATGTTATGGACATACATCTAATTATTATTAAGAGGAAATTGTTGAAAGTTAACGTTTAA
Protein
MSREVENILQSKIKYGIISIPMGSLDVFNKSRNVEYFEGAKDNLPDNSAQNTALKIKNLITQLNKDDLLLVLISGGGSALLPLPKSPITLEEKIGLVKKLANSGADIKELNTVRKVISDLKGGQLAVKAQPAQVVSLILSDIVGDPLDLIASGPTVQNTDGANKAVDVLKKYNLIDALPKSVQTLLENNGDNLVFPTNNTSNYIIGSNKISTKAAVVQCIELNYLPLVLSNKVTGNVQDVANKYSKLVTVICKYLRQNLEIDELKSNIKKLEISGTDLKVLNEIKISNKKPLCLILGGEITVAVKGTGKGGRNQQLALEFSKYLHKVKDQLNDFDIFILSAGTDGIDGPTDAAGAIGYLNLISESTADGLDVDKYLANNDTYNFFKLFKNDHLHVFTGHTNTNVMDIHLIIIKRKLLKVNV
Summary
Uniprot
H9IU97
A0A194QE11
A0A0N0PEH9
A0A2W1BF86
A0A2H1VSQ8
A0A0L7LHV0
+ More
A0A212FA60 A0A091QWP2 A0A2I0TMX8 U3K2M6 A0A226NQI7 G1KFJ3 A0A091MV60 A0A3M0JG41 A0A091KY42 A0A226MS82 A0A2P8XM22 A0A2D4IHM7 A0A087VJA8 V8PI43 A0A093EUP7 A0A091IZC0 A0A093GXC5 A0A218VCS1 A0A091NRM2 G1PRX0 G1MWH3 A0A2P4SY99 F6QH72 L5K804 V4APZ2 A0A091EN18 A0A091DR29 A0A094MMM6 A0A091PIZ9 A0A093IV18 A0A091H809 A0A087QTA2 H0Z6U8 A0A3Q0H2D6 A0A091R1N0 A0A3Q2U5V7 A0A1B6I196 A0A1B6JLL0 A0A091NYI7 A0A3Q7RAY1 K9IL55 A0A2F0BIF4 A0A093Q3B9 A0A3L7HP55 A0A383ZHZ8 A0A0Q3MPZ7 G5AXM5 R0JHG7 A0A091VC28 W5P6K4 R4WJW7 A0A2Y9J061 U3IVB9 A0A286XPC0 A0A3Q7VW24 A0A384D556 A0A2K5QF05 F6UZ82 A0A2Y9R4I5 A0A096MYQ5 A0A2J7PHR9 H3DBB7 A0A384D4C7 A0A067RI67 A0A3Q7W642 A0A341BHJ1 A0A2Y9N364 A0A093FL79 V9KL14 A0A2K5MCF9 A0A0D9RMT0 A0A340WSQ6 M3YJN2 A0A2K6TVF0 A0A1A6H8V1 D2A244 A0A2K5Y1R1 F1SIX9 G1R637 F7IB83 A0A2K6CP42 A0A1U7T2N0 A0A0R4J1H2 G7MV99 G7NZW3 A0A2K5V6G9 G3GZJ2 A0A1U8BZA1 M3WN46 A0A3Q7Q640 A0A2I0LSL6 A0A2U3WQP4 A0A2K5E0P4 A0A2K6M859
A0A212FA60 A0A091QWP2 A0A2I0TMX8 U3K2M6 A0A226NQI7 G1KFJ3 A0A091MV60 A0A3M0JG41 A0A091KY42 A0A226MS82 A0A2P8XM22 A0A2D4IHM7 A0A087VJA8 V8PI43 A0A093EUP7 A0A091IZC0 A0A093GXC5 A0A218VCS1 A0A091NRM2 G1PRX0 G1MWH3 A0A2P4SY99 F6QH72 L5K804 V4APZ2 A0A091EN18 A0A091DR29 A0A094MMM6 A0A091PIZ9 A0A093IV18 A0A091H809 A0A087QTA2 H0Z6U8 A0A3Q0H2D6 A0A091R1N0 A0A3Q2U5V7 A0A1B6I196 A0A1B6JLL0 A0A091NYI7 A0A3Q7RAY1 K9IL55 A0A2F0BIF4 A0A093Q3B9 A0A3L7HP55 A0A383ZHZ8 A0A0Q3MPZ7 G5AXM5 R0JHG7 A0A091VC28 W5P6K4 R4WJW7 A0A2Y9J061 U3IVB9 A0A286XPC0 A0A3Q7VW24 A0A384D556 A0A2K5QF05 F6UZ82 A0A2Y9R4I5 A0A096MYQ5 A0A2J7PHR9 H3DBB7 A0A384D4C7 A0A067RI67 A0A3Q7W642 A0A341BHJ1 A0A2Y9N364 A0A093FL79 V9KL14 A0A2K5MCF9 A0A0D9RMT0 A0A340WSQ6 M3YJN2 A0A2K6TVF0 A0A1A6H8V1 D2A244 A0A2K5Y1R1 F1SIX9 G1R637 F7IB83 A0A2K6CP42 A0A1U7T2N0 A0A0R4J1H2 G7MV99 G7NZW3 A0A2K5V6G9 G3GZJ2 A0A1U8BZA1 M3WN46 A0A3Q7Q640 A0A2I0LSL6 A0A2U3WQP4 A0A2K5E0P4 A0A2K6M859
Pubmed
19121390
26354079
28756777
26227816
22118469
24621616
+ More
29403074 24297900 21993624 20838655 19892987 23258410 23254933 20360741 15592404 29704459 21993625 20809919 23691247 24813606 17495919 15496914 24845553 24402279 18362917 19820115 19468303 21183079 23576753 22002653 25319552 21804562 17975172 23371554
29403074 24297900 21993624 20838655 19892987 23258410 23254933 20360741 15592404 29704459 21993625 20809919 23691247 24813606 17495919 15496914 24845553 24402279 18362917 19820115 19468303 21183079 23576753 22002653 25319552 21804562 17975172 23371554
EMBL
BABH01041648
KQ459193
KPJ03185.1
KQ459941
KPJ19173.1
KZ150086
+ More
PZC73769.1 ODYU01004216 SOQ43857.1 JTDY01001009 KOB75133.1 AGBW02009523 OWR50599.1 KK805269 KFQ31648.1 KZ508476 PKU35154.1 AGTO01011004 AWGT02003816 OXB69741.1 KK517875 KFP65375.1 QRBI01000145 RMC00002.1 KK761267 KFP45012.1 MCFN01000491 OXB58141.1 PYGN01001740 PSN33057.1 IACK01105309 LAA83731.1 KL495507 KFO12700.1 AZIM01000063 ETE73641.1 KL469282 KFV18221.1 KL218426 KFP04886.1 KL216884 KFV71754.1 MUZQ01000011 OWK63648.1 KL391382 KFP91732.1 AAPE02022320 PPHD01016616 POI29102.1 KB030947 ELK07839.1 KB201037 ESO99287.1 KK718710 KFO57987.1 KN122133 KFO32695.1 KL345428 KFZ55498.1 KK674021 KFQ07341.1 KK601669 KFW06550.1 KL448312 KFO82382.1 KL225895 KFM04456.1 ABQF01031688 KK711528 KFQ33690.1 AADN05000356 GECU01027052 JAS80654.1 GECU01007582 JAT00125.1 KK647522 KFP99800.1 GABZ01005537 JAA47988.1 NTJE010219635 MBW02779.1 KL670312 KFW78697.1 RAZU01000165 RLQ67643.1 LMAW01001069 KQK84597.1 JH167404 GEBF01005062 EHB01786.1 JAN98570.1 KB744014 EOA96391.1 KK423628 KFQ87213.1 AMGL01045544 AMGL01045545 AK417910 BAN21125.1 AAKN02012961 AHZZ02020093 NEVH01025133 PNF15868.1 KK852460 KDR23492.1 KK629652 KFV55049.1 JW866364 AFO98881.1 AQIB01131957 AEYP01099203 LZPO01044192 OBS75013.1 KQ971338 EFA02735.1 AEMK02000086 ADFV01173422 AC164430 JU322921 CM001258 AFE66677.1 EHH26660.1 CM001277 EHH51266.1 AQIA01039752 JH000078 EGW05667.1 AANG04000016 AKCR02000111 PKK20419.1
PZC73769.1 ODYU01004216 SOQ43857.1 JTDY01001009 KOB75133.1 AGBW02009523 OWR50599.1 KK805269 KFQ31648.1 KZ508476 PKU35154.1 AGTO01011004 AWGT02003816 OXB69741.1 KK517875 KFP65375.1 QRBI01000145 RMC00002.1 KK761267 KFP45012.1 MCFN01000491 OXB58141.1 PYGN01001740 PSN33057.1 IACK01105309 LAA83731.1 KL495507 KFO12700.1 AZIM01000063 ETE73641.1 KL469282 KFV18221.1 KL218426 KFP04886.1 KL216884 KFV71754.1 MUZQ01000011 OWK63648.1 KL391382 KFP91732.1 AAPE02022320 PPHD01016616 POI29102.1 KB030947 ELK07839.1 KB201037 ESO99287.1 KK718710 KFO57987.1 KN122133 KFO32695.1 KL345428 KFZ55498.1 KK674021 KFQ07341.1 KK601669 KFW06550.1 KL448312 KFO82382.1 KL225895 KFM04456.1 ABQF01031688 KK711528 KFQ33690.1 AADN05000356 GECU01027052 JAS80654.1 GECU01007582 JAT00125.1 KK647522 KFP99800.1 GABZ01005537 JAA47988.1 NTJE010219635 MBW02779.1 KL670312 KFW78697.1 RAZU01000165 RLQ67643.1 LMAW01001069 KQK84597.1 JH167404 GEBF01005062 EHB01786.1 JAN98570.1 KB744014 EOA96391.1 KK423628 KFQ87213.1 AMGL01045544 AMGL01045545 AK417910 BAN21125.1 AAKN02012961 AHZZ02020093 NEVH01025133 PNF15868.1 KK852460 KDR23492.1 KK629652 KFV55049.1 JW866364 AFO98881.1 AQIB01131957 AEYP01099203 LZPO01044192 OBS75013.1 KQ971338 EFA02735.1 AEMK02000086 ADFV01173422 AC164430 JU322921 CM001258 AFE66677.1 EHH26660.1 CM001277 EHH51266.1 AQIA01039752 JH000078 EGW05667.1 AANG04000016 AKCR02000111 PKK20419.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000016665
+ More
UP000198419 UP000001646 UP000269221 UP000198323 UP000245037 UP000054308 UP000053875 UP000197619 UP000001074 UP000001645 UP000002281 UP000010552 UP000030746 UP000052976 UP000028990 UP000053760 UP000053286 UP000007754 UP000189705 UP000000539 UP000286640 UP000053258 UP000273346 UP000261681 UP000051836 UP000006813 UP000002356 UP000248482 UP000005447 UP000286642 UP000261680 UP000291021 UP000233040 UP000002280 UP000248480 UP000028761 UP000235965 UP000007303 UP000027135 UP000252040 UP000248483 UP000233060 UP000029965 UP000265300 UP000000715 UP000233220 UP000092124 UP000007266 UP000233140 UP000008227 UP000001073 UP000008225 UP000233120 UP000189704 UP000000589 UP000009130 UP000233100 UP000001075 UP000189706 UP000011712 UP000286641 UP000053872 UP000245340 UP000233020 UP000233180
UP000198419 UP000001646 UP000269221 UP000198323 UP000245037 UP000054308 UP000053875 UP000197619 UP000001074 UP000001645 UP000002281 UP000010552 UP000030746 UP000052976 UP000028990 UP000053760 UP000053286 UP000007754 UP000189705 UP000000539 UP000286640 UP000053258 UP000273346 UP000261681 UP000051836 UP000006813 UP000002356 UP000248482 UP000005447 UP000286642 UP000261680 UP000291021 UP000233040 UP000002280 UP000248480 UP000028761 UP000235965 UP000007303 UP000027135 UP000252040 UP000248483 UP000233060 UP000029965 UP000265300 UP000000715 UP000233220 UP000092124 UP000007266 UP000233140 UP000008227 UP000001073 UP000008225 UP000233120 UP000189704 UP000000589 UP000009130 UP000233100 UP000001075 UP000189706 UP000011712 UP000286641 UP000053872 UP000245340 UP000233020 UP000233180
Interpro
Gene 3D
ProteinModelPortal
H9IU97
A0A194QE11
A0A0N0PEH9
A0A2W1BF86
A0A2H1VSQ8
A0A0L7LHV0
+ More
A0A212FA60 A0A091QWP2 A0A2I0TMX8 U3K2M6 A0A226NQI7 G1KFJ3 A0A091MV60 A0A3M0JG41 A0A091KY42 A0A226MS82 A0A2P8XM22 A0A2D4IHM7 A0A087VJA8 V8PI43 A0A093EUP7 A0A091IZC0 A0A093GXC5 A0A218VCS1 A0A091NRM2 G1PRX0 G1MWH3 A0A2P4SY99 F6QH72 L5K804 V4APZ2 A0A091EN18 A0A091DR29 A0A094MMM6 A0A091PIZ9 A0A093IV18 A0A091H809 A0A087QTA2 H0Z6U8 A0A3Q0H2D6 A0A091R1N0 A0A3Q2U5V7 A0A1B6I196 A0A1B6JLL0 A0A091NYI7 A0A3Q7RAY1 K9IL55 A0A2F0BIF4 A0A093Q3B9 A0A3L7HP55 A0A383ZHZ8 A0A0Q3MPZ7 G5AXM5 R0JHG7 A0A091VC28 W5P6K4 R4WJW7 A0A2Y9J061 U3IVB9 A0A286XPC0 A0A3Q7VW24 A0A384D556 A0A2K5QF05 F6UZ82 A0A2Y9R4I5 A0A096MYQ5 A0A2J7PHR9 H3DBB7 A0A384D4C7 A0A067RI67 A0A3Q7W642 A0A341BHJ1 A0A2Y9N364 A0A093FL79 V9KL14 A0A2K5MCF9 A0A0D9RMT0 A0A340WSQ6 M3YJN2 A0A2K6TVF0 A0A1A6H8V1 D2A244 A0A2K5Y1R1 F1SIX9 G1R637 F7IB83 A0A2K6CP42 A0A1U7T2N0 A0A0R4J1H2 G7MV99 G7NZW3 A0A2K5V6G9 G3GZJ2 A0A1U8BZA1 M3WN46 A0A3Q7Q640 A0A2I0LSL6 A0A2U3WQP4 A0A2K5E0P4 A0A2K6M859
A0A212FA60 A0A091QWP2 A0A2I0TMX8 U3K2M6 A0A226NQI7 G1KFJ3 A0A091MV60 A0A3M0JG41 A0A091KY42 A0A226MS82 A0A2P8XM22 A0A2D4IHM7 A0A087VJA8 V8PI43 A0A093EUP7 A0A091IZC0 A0A093GXC5 A0A218VCS1 A0A091NRM2 G1PRX0 G1MWH3 A0A2P4SY99 F6QH72 L5K804 V4APZ2 A0A091EN18 A0A091DR29 A0A094MMM6 A0A091PIZ9 A0A093IV18 A0A091H809 A0A087QTA2 H0Z6U8 A0A3Q0H2D6 A0A091R1N0 A0A3Q2U5V7 A0A1B6I196 A0A1B6JLL0 A0A091NYI7 A0A3Q7RAY1 K9IL55 A0A2F0BIF4 A0A093Q3B9 A0A3L7HP55 A0A383ZHZ8 A0A0Q3MPZ7 G5AXM5 R0JHG7 A0A091VC28 W5P6K4 R4WJW7 A0A2Y9J061 U3IVB9 A0A286XPC0 A0A3Q7VW24 A0A384D556 A0A2K5QF05 F6UZ82 A0A2Y9R4I5 A0A096MYQ5 A0A2J7PHR9 H3DBB7 A0A384D4C7 A0A067RI67 A0A3Q7W642 A0A341BHJ1 A0A2Y9N364 A0A093FL79 V9KL14 A0A2K5MCF9 A0A0D9RMT0 A0A340WSQ6 M3YJN2 A0A2K6TVF0 A0A1A6H8V1 D2A244 A0A2K5Y1R1 F1SIX9 G1R637 F7IB83 A0A2K6CP42 A0A1U7T2N0 A0A0R4J1H2 G7MV99 G7NZW3 A0A2K5V6G9 G3GZJ2 A0A1U8BZA1 M3WN46 A0A3Q7Q640 A0A2I0LSL6 A0A2U3WQP4 A0A2K5E0P4 A0A2K6M859
PDB
2B8N
E-value=4.05574e-47,
Score=475
Ontologies
PATHWAY
00030
Pentose phosphate pathway - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00561 Glycerolipid metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
PANTHER
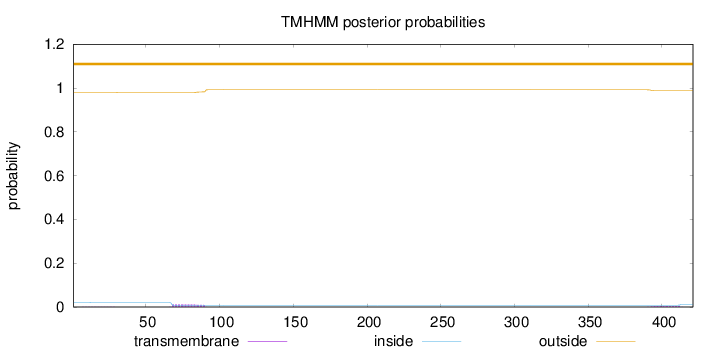
Topology
Length:
421
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.360360000000001
Exp number, first 60 AAs:
0.01174
Total prob of N-in:
0.02058
outside
1 - 421
Population Genetic Test Statistics
Pi
174.472068
Theta
183.514552
Tajima's D
0.174278
CLR
21.425807
CSRT
0.418029098545073
Interpretation
Uncertain
