Pre Gene Modal
BGIBMGA000828
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_DNA_replication_licensing_factor_Mcm6-like_[Bombyx_mori]
Full name
DNA helicase
+ More
DNA replication licensing factor Mcm6
DNA replication licensing factor Mcm6
Location in the cell
Cytoplasmic Reliability : 2.562
Sequence
CDS
ATGGATGTAGCTGACACATATGCCACACAGAGTCAAGTAAAAGATGAAGTGGGAATTCGATGCCAAAAGCTTTTTCAAGACTTCTTAGAAGAATTTAAGGAAGATAACGAAATCAAATATGAGAAACATGCTAAGGAACTATTGAAACCAGAGCTCAGTACCCTTGAAGTTAGTTTTGATGATGTTGAAAAGTACAACCAGAATCTTGCTACCACAATAATTGAAGAGTATTATAGGATATATCCATTTCTAAACAGAGCCATTTTGAATTACATACTCAGTCTTGCGGAAACTGGAATGAAAAAAGATCTACAAGATAAAGAATGCTATGTTTCATTTGTTGATGTACCAACAAGACACAAAGTAAGGGAGTTAACCACAGCAAAGATTGGTACTCTCATAAGGATCTCTGGGCAGATTGTAAGGACACATCCAGTTCACCCTGAATTAGTTCAGGGCACATTTGTGTGCTTAGATTGTCAAACAGTTATTAAAAATGTAGAGCAACAGTTTAAGTACACAATTCCAACAATCTGCCGTAATCCTGTATGTGCTAATAGAAGGCGTTTTATGCTGGACGCAGACAAGAGCGTTTTTGTTGACTTCCAAAAGATACGAATCCAGGAAACCCAAGCTGAATTGCCCCGTGGTTGCATACCCAGATCTCTTGAAGTGATATTGAGGGCTGAAGCTGTTGAATCTGTGCAGGCTGGTGACCGTTATGATTTTACAGGAACACTGATTGTAGTGCCTGATGTGGGAGCCTTGTCAATGCCAGGCTCCAGGGCTGAGATTACAACCAGGACTAAATTAGCAAATGAAGGTCAAATGGAAGGCATAAAGGGACTCAAGGCTCTTGGAGTGCGAGAACTGCATTATAAAACAGCATTTTTAGCATGCAGTGTACAAGCTGTTTCTAGAAGGTTTGGTACTGCAGAGTTGCCTACACATGACCTAACAACTGAAGACATGAGGAAACAAATGACTGACAAAGAATGGGACAAGGTTTATGAAATGTCTCGAGATCGCAATTTATATAATAATTTAATCACGAGCTTGTTTCCAAGCATTCATGGAAATAATGAGGTGAAGCGAGGCGTCCTATTGATGTTGTTTGGAGGGGTCGCCAAAACTACTATTGAAGGAACAACACTTCGAGGCGACATAAACGTGTGCATCGTAGGAGACCCGAGTACAGCCAAGTCCCAGTTACTGAAGCAAGTCAGTGAGATCACACCCCGAGCCGTGTACACTAGCGGAAAGGCTTCCTCTGCTGCTGGTCTCACTGCAGCTGTTGTTAGGGACGAAGAATCCTTCGATTTCGTGATCGAAGCAGGCGCTTTAATGTTAGCTGACAACGGTGTGTGCTGTATTGACGAGTTCGACAAGATGGATCCTGGTGATCAAGTTGCAATACACGAGGCCATGGAACAACAAACAATTTCCTTGGCTAAGGCCGGAGTGAGGGCTACCCTAAATGCACGTACGTCGATACTTGCTGCCGCAAATCCTATTGGCGGACGATACGATCGAGCTAAATCCTTACAACAAAACGTCGCACTGAGCCCCCCCATTATGTCACGTTTCGATTTGTTCTTCATATTGATAGACGAGAGCAGTGAAATGGTTGATTACGCAATAGCAAGGAAAATTGTCGATCTGCACTGCAATAAAGAGGAGAGCTATGATTGTGTTTACTCAAGGGACGACCTGCTCAGGTACATTGCGTTCGCGAGGTCATTCAAGCCCGTCATTACCGAGGAGGCAGGCAAGTTACTGGTGGAGTACTACGCGTTGCTGCGGGGCCGTGAGGGGGGCGCGGGGGGCGGCGCGTGGCGCATCACGGTGCGCCAGCTCGAGTCCATGGTGCGGCTCGCCGAGGGCGTCGCCAAGATGCACTGCAGCGGACACGTCACGCCCGCGCACGTGCACGAGGCCTACCGATTACTAAATAAATCCATTATTCGCGTGGAGCAGCCCGATATACATTTGGACGAGGATGAACCTCAATGCGAGCCGAGCATGGATGTGGATCAAGACGAACCTAATGGCACTGCAGAAACTCCCAGCAATGGTGATTCGGCGCCTAAAAAGAAACTGGCGCTTTCATTCGAAGAATACAAAAGCCTCAGTAATATGCTGGTCGTGTACATGAGGAAGGAAGAGGCTGAAGCGGAGACGAGAGACGAGGAGAGCTCCGGTATGCACAAGTCTGCCGTAGTGAGCTGGTACCTAGAACAGCTGGTGGCACAAGGACAAATCGAAAGCGAAGACGAGTTACTTGAACGAAAGACACTAGTTGAGAAAGTCATTGATAGACTTATGTACCATGTCATAAATTCTGTCACATGTTTAATGTAA
Protein
MDVADTYATQSQVKDEVGIRCQKLFQDFLEEFKEDNEIKYEKHAKELLKPELSTLEVSFDDVEKYNQNLATTIIEEYYRIYPFLNRAILNYILSLAETGMKKDLQDKECYVSFVDVPTRHKVRELTTAKIGTLIRISGQIVRTHPVHPELVQGTFVCLDCQTVIKNVEQQFKYTIPTICRNPVCANRRRFMLDADKSVFVDFQKIRIQETQAELPRGCIPRSLEVILRAEAVESVQAGDRYDFTGTLIVVPDVGALSMPGSRAEITTRTKLANEGQMEGIKGLKALGVRELHYKTAFLACSVQAVSRRFGTAELPTHDLTTEDMRKQMTDKEWDKVYEMSRDRNLYNNLITSLFPSIHGNNEVKRGVLLMLFGGVAKTTIEGTTLRGDINVCIVGDPSTAKSQLLKQVSEITPRAVYTSGKASSAAGLTAAVVRDEESFDFVIEAGALMLADNGVCCIDEFDKMDPGDQVAIHEAMEQQTISLAKAGVRATLNARTSILAAANPIGGRYDRAKSLQQNVALSPPIMSRFDLFFILIDESSEMVDYAIARKIVDLHCNKEESYDCVYSRDDLLRYIAFARSFKPVITEEAGKLLVEYYALLRGREGGAGGGAWRITVRQLESMVRLAEGVAKMHCSGHVTPAHVHEAYRLLNKSIIRVEQPDIHLDEDEPQCEPSMDVDQDEPNGTAETPSNGDSAPKKKLALSFEEYKSLSNMLVVYMRKEEAEAETRDEESSGMHKSAVVSWYLEQLVAQGQIESEDELLERKTLVEKVIDRLMYHVINSVTCLM
Summary
Description
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity (By similarity).
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity Required for DNA replication and cell proliferation. Required for mitotic cycles, endocycles, and the special S phase associated with the amplification of chorion genes; has a role in origin unwinding or fork elongation at chorion loci (By similarity).
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity Required for DNA replication and cell proliferation. Required for mitotic cycles, endocycles, and the special S phase associated with the amplification of chorion genes; has a role in origin unwinding or fork elongation at chorion loci.
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity Required for DNA replication and cell proliferation. Required for mitotic cycles, endocycles, and the special S phase associated with the amplification of chorion genes; has a role in origin unwinding or fork elongation at chorion loci (By similarity).
Acts as component of the Mcm2-7 complex (Mcm complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the Mcm2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity Required for DNA replication and cell proliferation. Required for mitotic cycles, endocycles, and the special S phase associated with the amplification of chorion genes; has a role in origin unwinding or fork elongation at chorion loci.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5 (By simililarity). The heterodimers of Mcm4/Mcm6 and Mcm3/Mcm5 interact with Mcm2 and Mcm7 (By similarity).
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5. The heterodimers of mcm4/mcm6 and mcm3/mcm5 interact with mcm2 and mcm7 (By similarity).
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5 (Probable). The heterodimers of Mcm4/Mcm6 and Mcm3/Mcm5 interact with Mcm2 and Mcm7.
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5. The heterodimers of mcm4/mcm6 and mcm3/mcm5 interact with mcm2 and mcm7 (By similarity).
Component of the Mcm2-7 complex. The complex forms a toroidal hexameric ring with the proposed subunit order Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5 (Probable). The heterodimers of Mcm4/Mcm6 and Mcm3/Mcm5 interact with Mcm2 and Mcm7.
Similarity
Belongs to the MCM family.
Keywords
ATP-binding
Cell cycle
Cell division
Complete proteome
DNA replication
DNA-binding
Helicase
Hydrolase
Metal-binding
Mitosis
Nucleotide-binding
Nucleus
Reference proteome
Zinc
Zinc-finger
Feature
chain DNA replication licensing factor Mcm6
Uniprot
H9IU98
A0A2H1VSP1
A0A2W1BKG5
A0A3S2TK09
A0A212FA39
A0A194QCB2
+ More
A0A2A4J5H3 B0WPF1 A0A1Q3EVC8 A0A182FMV2 A0A2J7RDP5 A0A182HC06 A0A084WD26 W5JLP0 A0A182J130 A0A067RF20 A0A182N8Q8 A0A182QHU7 A0A336LRZ0 A0A182R6W1 A0A182MQ41 A0A182PV53 A0A182VSY2 Q16LS4 A0A2P8XKU6 A0A182XB67 A0A182KJS4 A0A1I8NVM4 A0A182I0R1 A0A182URP9 Q7Q0Q1 A0A0A1WQ29 A0A182JPH2 A0A034VW54 A0A1W4XIQ3 W8B565 T1P8H7 A0A1A9WKB8 A0A089HMH5 A0A0K8V2U0 A0A182YHC6 A0A1B0CE05 A0A0L0CNC4 B3MS81 A0A1A9ZNZ9 B4L5X6 A0A1A9VQ30 A0A1A9Y544 B4JMY9 A0A0M4FAP2 Q29JI9 D0QWF4 A0A0C9PH07 B4NPC9 A0A3B0JF20 B4PYU3 N6T4M5 A0A1W4VEJ0 B3NXU3 B4M7G8 B4R5N4 C6SUY3 Q9V461 D6X3S8 A0A1B6J7E9 B4I083 K7IXP3 U4UD49 A0A182TR97 A0A1B6ECS9 A0A1J1I3Q6 A0A087ZNB8 A0A154PRR2 A0A0J7L6G6 A0A146M089 A0A0A9X784 A0A2A3E600 A0A026WMD9 A0A023F4Y8 A0A151X131 V4BFH9 A0A2R7WPJ4 A0A1B0BXW1 A0A151I153 T1I9N9 A0A158NMR3 F4WN06 E2BEY5 A0A151IBU6 A0A151JVN1 A0A1Y1M0A5 E2ALN2 E9J2F5 A0A1B0D5I2 A0A1S3K5V8 A0A1S3I664 A0A310SE60 A0A0B7B3B6 A0A0L7R4F7 A0A0B7AZT6
A0A2A4J5H3 B0WPF1 A0A1Q3EVC8 A0A182FMV2 A0A2J7RDP5 A0A182HC06 A0A084WD26 W5JLP0 A0A182J130 A0A067RF20 A0A182N8Q8 A0A182QHU7 A0A336LRZ0 A0A182R6W1 A0A182MQ41 A0A182PV53 A0A182VSY2 Q16LS4 A0A2P8XKU6 A0A182XB67 A0A182KJS4 A0A1I8NVM4 A0A182I0R1 A0A182URP9 Q7Q0Q1 A0A0A1WQ29 A0A182JPH2 A0A034VW54 A0A1W4XIQ3 W8B565 T1P8H7 A0A1A9WKB8 A0A089HMH5 A0A0K8V2U0 A0A182YHC6 A0A1B0CE05 A0A0L0CNC4 B3MS81 A0A1A9ZNZ9 B4L5X6 A0A1A9VQ30 A0A1A9Y544 B4JMY9 A0A0M4FAP2 Q29JI9 D0QWF4 A0A0C9PH07 B4NPC9 A0A3B0JF20 B4PYU3 N6T4M5 A0A1W4VEJ0 B3NXU3 B4M7G8 B4R5N4 C6SUY3 Q9V461 D6X3S8 A0A1B6J7E9 B4I083 K7IXP3 U4UD49 A0A182TR97 A0A1B6ECS9 A0A1J1I3Q6 A0A087ZNB8 A0A154PRR2 A0A0J7L6G6 A0A146M089 A0A0A9X784 A0A2A3E600 A0A026WMD9 A0A023F4Y8 A0A151X131 V4BFH9 A0A2R7WPJ4 A0A1B0BXW1 A0A151I153 T1I9N9 A0A158NMR3 F4WN06 E2BEY5 A0A151IBU6 A0A151JVN1 A0A1Y1M0A5 E2ALN2 E9J2F5 A0A1B0D5I2 A0A1S3K5V8 A0A1S3I664 A0A310SE60 A0A0B7B3B6 A0A0L7R4F7 A0A0B7AZT6
EC Number
3.6.4.12
Pubmed
19121390
28756777
22118469
26354079
26483478
24438588
+ More
20920257 23761445 24845553 17510324 29403074 20966253 12364791 25830018 25348373 24495485 25315136 25244985 26108605 17994087 15632085 19859648 17550304 23537049 9795205 10023044 11854416 10731132 12537572 12537569 16798881 20122406 18362917 19820115 20075255 26823975 25401762 24508170 30249741 25474469 23254933 21347285 21719571 20798317 28004739 21282665
20920257 23761445 24845553 17510324 29403074 20966253 12364791 25830018 25348373 24495485 25315136 25244985 26108605 17994087 15632085 19859648 17550304 23537049 9795205 10023044 11854416 10731132 12537572 12537569 16798881 20122406 18362917 19820115 20075255 26823975 25401762 24508170 30249741 25474469 23254933 21347285 21719571 20798317 28004739 21282665
EMBL
BABH01041643
BABH01041644
BABH01041645
BABH01041646
ODYU01004216
SOQ43855.1
+ More
KZ150086 PZC73767.1 RSAL01000087 RVE48249.1 AGBW02009523 OWR50601.1 KQ459193 KPJ03183.1 NWSH01002958 PCG67221.1 DS232023 EDS32330.1 GFDL01015785 JAV19260.1 NEVH01005287 PNF38953.1 JXUM01032592 KQ560961 KXJ80194.1 ATLV01022923 KE525338 KFB48120.1 ADMH02001180 ETN63820.1 KK852676 KDR18687.1 AXCN02001859 UFQT01000086 SSX19493.1 AXCM01000229 CH477894 EAT35274.1 PYGN01001830 PSN32621.1 APCN01005234 AAAB01008980 GBXI01013769 JAD00523.1 GAKP01011381 GAKP01011380 JAC47571.1 GAMC01014317 JAB92238.1 KA644959 AFP59588.1 KM036514 AIP98401.1 GDHF01019409 JAI32905.1 AJWK01008460 JRES01000147 KNC33820.1 CH902622 EDV34636.1 CH933811 EDW06585.1 CH916371 EDV92082.1 CP012528 ALC49507.1 CH379063 EAL32312.1 FJ821027 ACN94664.1 GBYB01000143 JAG69910.1 CH964291 EDW86369.1 OUUW01000003 SPP78802.1 CM000162 EDX01029.1 APGK01044211 KB741021 ENN75064.1 CH954180 EDV47394.1 CH940653 EDW62735.1 CM000366 EDX17271.1 BT088975 ACT82957.1 AB010108 AF124744 AE014298 AY052102 AAD32858.1 AAF46184.1 AAK93526.1 BAA34732.1 KQ971326 EEZ97375.1 GECU01012609 JAS95097.1 CH480819 EDW52914.1 KB632003 ERL87840.1 GEDC01016650 GEDC01001601 JAS20648.1 JAS35697.1 CVRI01000038 CRK94228.1 KQ435031 KZC14034.1 LBMM01000485 KMQ98246.1 GDHC01005371 JAQ13258.1 GBHO01028961 JAG14643.1 KZ288357 PBC27135.1 KK107152 QOIP01000007 EZA57232.1 RLU20550.1 GBBI01002609 JAC16103.1 KQ982588 KYQ54107.1 KB202849 ESO87684.1 KK855225 PTY21523.1 JXJN01022373 JXJN01022374 KQ976593 KYM79671.1 ACPB03017705 ADTU01020709 GL888225 EGI64419.1 GL447885 EFN85750.1 KQ978078 KYM97194.1 KQ981689 KYN37892.1 GEZM01045606 JAV77725.1 GL440609 EFN65641.1 GL767845 EFZ12998.1 AJVK01025284 KQ760761 OAD59139.1 HACG01039745 CEK86610.1 KQ414657 KOC65762.1 HACG01039743 HACG01039744 CEK86608.1 CEK86609.1
KZ150086 PZC73767.1 RSAL01000087 RVE48249.1 AGBW02009523 OWR50601.1 KQ459193 KPJ03183.1 NWSH01002958 PCG67221.1 DS232023 EDS32330.1 GFDL01015785 JAV19260.1 NEVH01005287 PNF38953.1 JXUM01032592 KQ560961 KXJ80194.1 ATLV01022923 KE525338 KFB48120.1 ADMH02001180 ETN63820.1 KK852676 KDR18687.1 AXCN02001859 UFQT01000086 SSX19493.1 AXCM01000229 CH477894 EAT35274.1 PYGN01001830 PSN32621.1 APCN01005234 AAAB01008980 GBXI01013769 JAD00523.1 GAKP01011381 GAKP01011380 JAC47571.1 GAMC01014317 JAB92238.1 KA644959 AFP59588.1 KM036514 AIP98401.1 GDHF01019409 JAI32905.1 AJWK01008460 JRES01000147 KNC33820.1 CH902622 EDV34636.1 CH933811 EDW06585.1 CH916371 EDV92082.1 CP012528 ALC49507.1 CH379063 EAL32312.1 FJ821027 ACN94664.1 GBYB01000143 JAG69910.1 CH964291 EDW86369.1 OUUW01000003 SPP78802.1 CM000162 EDX01029.1 APGK01044211 KB741021 ENN75064.1 CH954180 EDV47394.1 CH940653 EDW62735.1 CM000366 EDX17271.1 BT088975 ACT82957.1 AB010108 AF124744 AE014298 AY052102 AAD32858.1 AAF46184.1 AAK93526.1 BAA34732.1 KQ971326 EEZ97375.1 GECU01012609 JAS95097.1 CH480819 EDW52914.1 KB632003 ERL87840.1 GEDC01016650 GEDC01001601 JAS20648.1 JAS35697.1 CVRI01000038 CRK94228.1 KQ435031 KZC14034.1 LBMM01000485 KMQ98246.1 GDHC01005371 JAQ13258.1 GBHO01028961 JAG14643.1 KZ288357 PBC27135.1 KK107152 QOIP01000007 EZA57232.1 RLU20550.1 GBBI01002609 JAC16103.1 KQ982588 KYQ54107.1 KB202849 ESO87684.1 KK855225 PTY21523.1 JXJN01022373 JXJN01022374 KQ976593 KYM79671.1 ACPB03017705 ADTU01020709 GL888225 EGI64419.1 GL447885 EFN85750.1 KQ978078 KYM97194.1 KQ981689 KYN37892.1 GEZM01045606 JAV77725.1 GL440609 EFN65641.1 GL767845 EFZ12998.1 AJVK01025284 KQ760761 OAD59139.1 HACG01039745 CEK86610.1 KQ414657 KOC65762.1 HACG01039743 HACG01039744 CEK86608.1 CEK86609.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000218220
UP000002320
+ More
UP000069272 UP000235965 UP000069940 UP000249989 UP000030765 UP000000673 UP000075880 UP000027135 UP000075884 UP000075886 UP000075900 UP000075883 UP000075885 UP000075920 UP000008820 UP000245037 UP000076407 UP000075882 UP000095300 UP000075840 UP000075903 UP000007062 UP000075881 UP000192223 UP000095301 UP000091820 UP000076408 UP000092461 UP000037069 UP000007801 UP000092445 UP000009192 UP000078200 UP000092443 UP000001070 UP000092553 UP000001819 UP000007798 UP000268350 UP000002282 UP000019118 UP000192221 UP000008711 UP000008792 UP000000304 UP000000803 UP000007266 UP000001292 UP000002358 UP000030742 UP000075902 UP000183832 UP000005203 UP000076502 UP000036403 UP000242457 UP000053097 UP000279307 UP000075809 UP000030746 UP000092460 UP000078540 UP000015103 UP000005205 UP000007755 UP000008237 UP000078542 UP000078541 UP000000311 UP000092462 UP000085678 UP000053825
UP000069272 UP000235965 UP000069940 UP000249989 UP000030765 UP000000673 UP000075880 UP000027135 UP000075884 UP000075886 UP000075900 UP000075883 UP000075885 UP000075920 UP000008820 UP000245037 UP000076407 UP000075882 UP000095300 UP000075840 UP000075903 UP000007062 UP000075881 UP000192223 UP000095301 UP000091820 UP000076408 UP000092461 UP000037069 UP000007801 UP000092445 UP000009192 UP000078200 UP000092443 UP000001070 UP000092553 UP000001819 UP000007798 UP000268350 UP000002282 UP000019118 UP000192221 UP000008711 UP000008792 UP000000304 UP000000803 UP000007266 UP000001292 UP000002358 UP000030742 UP000075902 UP000183832 UP000005203 UP000076502 UP000036403 UP000242457 UP000053097 UP000279307 UP000075809 UP000030746 UP000092460 UP000078540 UP000015103 UP000005205 UP000007755 UP000008237 UP000078542 UP000078541 UP000000311 UP000092462 UP000085678 UP000053825
Interpro
Gene 3D
ProteinModelPortal
H9IU98
A0A2H1VSP1
A0A2W1BKG5
A0A3S2TK09
A0A212FA39
A0A194QCB2
+ More
A0A2A4J5H3 B0WPF1 A0A1Q3EVC8 A0A182FMV2 A0A2J7RDP5 A0A182HC06 A0A084WD26 W5JLP0 A0A182J130 A0A067RF20 A0A182N8Q8 A0A182QHU7 A0A336LRZ0 A0A182R6W1 A0A182MQ41 A0A182PV53 A0A182VSY2 Q16LS4 A0A2P8XKU6 A0A182XB67 A0A182KJS4 A0A1I8NVM4 A0A182I0R1 A0A182URP9 Q7Q0Q1 A0A0A1WQ29 A0A182JPH2 A0A034VW54 A0A1W4XIQ3 W8B565 T1P8H7 A0A1A9WKB8 A0A089HMH5 A0A0K8V2U0 A0A182YHC6 A0A1B0CE05 A0A0L0CNC4 B3MS81 A0A1A9ZNZ9 B4L5X6 A0A1A9VQ30 A0A1A9Y544 B4JMY9 A0A0M4FAP2 Q29JI9 D0QWF4 A0A0C9PH07 B4NPC9 A0A3B0JF20 B4PYU3 N6T4M5 A0A1W4VEJ0 B3NXU3 B4M7G8 B4R5N4 C6SUY3 Q9V461 D6X3S8 A0A1B6J7E9 B4I083 K7IXP3 U4UD49 A0A182TR97 A0A1B6ECS9 A0A1J1I3Q6 A0A087ZNB8 A0A154PRR2 A0A0J7L6G6 A0A146M089 A0A0A9X784 A0A2A3E600 A0A026WMD9 A0A023F4Y8 A0A151X131 V4BFH9 A0A2R7WPJ4 A0A1B0BXW1 A0A151I153 T1I9N9 A0A158NMR3 F4WN06 E2BEY5 A0A151IBU6 A0A151JVN1 A0A1Y1M0A5 E2ALN2 E9J2F5 A0A1B0D5I2 A0A1S3K5V8 A0A1S3I664 A0A310SE60 A0A0B7B3B6 A0A0L7R4F7 A0A0B7AZT6
A0A2A4J5H3 B0WPF1 A0A1Q3EVC8 A0A182FMV2 A0A2J7RDP5 A0A182HC06 A0A084WD26 W5JLP0 A0A182J130 A0A067RF20 A0A182N8Q8 A0A182QHU7 A0A336LRZ0 A0A182R6W1 A0A182MQ41 A0A182PV53 A0A182VSY2 Q16LS4 A0A2P8XKU6 A0A182XB67 A0A182KJS4 A0A1I8NVM4 A0A182I0R1 A0A182URP9 Q7Q0Q1 A0A0A1WQ29 A0A182JPH2 A0A034VW54 A0A1W4XIQ3 W8B565 T1P8H7 A0A1A9WKB8 A0A089HMH5 A0A0K8V2U0 A0A182YHC6 A0A1B0CE05 A0A0L0CNC4 B3MS81 A0A1A9ZNZ9 B4L5X6 A0A1A9VQ30 A0A1A9Y544 B4JMY9 A0A0M4FAP2 Q29JI9 D0QWF4 A0A0C9PH07 B4NPC9 A0A3B0JF20 B4PYU3 N6T4M5 A0A1W4VEJ0 B3NXU3 B4M7G8 B4R5N4 C6SUY3 Q9V461 D6X3S8 A0A1B6J7E9 B4I083 K7IXP3 U4UD49 A0A182TR97 A0A1B6ECS9 A0A1J1I3Q6 A0A087ZNB8 A0A154PRR2 A0A0J7L6G6 A0A146M089 A0A0A9X784 A0A2A3E600 A0A026WMD9 A0A023F4Y8 A0A151X131 V4BFH9 A0A2R7WPJ4 A0A1B0BXW1 A0A151I153 T1I9N9 A0A158NMR3 F4WN06 E2BEY5 A0A151IBU6 A0A151JVN1 A0A1Y1M0A5 E2ALN2 E9J2F5 A0A1B0D5I2 A0A1S3K5V8 A0A1S3I664 A0A310SE60 A0A0B7B3B6 A0A0L7R4F7 A0A0B7AZT6
PDB
6HV9
E-value=1.0644e-172,
Score=1560
Ontologies
GO
PANTHER
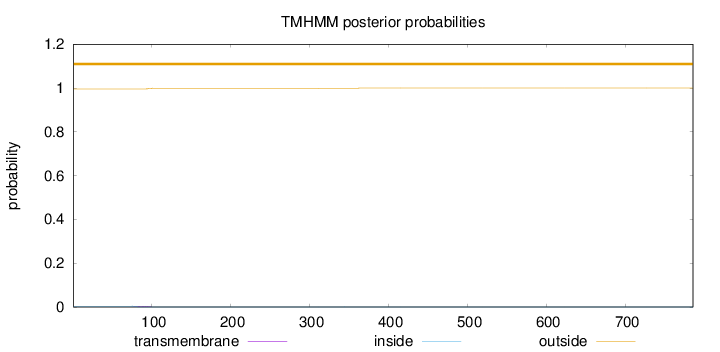
Topology
Subcellular location
Nucleus
Length:
786
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0702199999999997
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00386
outside
1 - 786
Population Genetic Test Statistics
Pi
198.19032
Theta
171.073044
Tajima's D
0.751162
CLR
0.038683
CSRT
0.587520623968802
Interpretation
Uncertain
