Gene
KWMTBOMO15656
Annotation
PREDICTED:_DNA_mismatch_repair_protein_Mlh1_isoform_X1_[Amyelois_transitella]
Full name
DNA mismatch repair protein Mlh1
Alternative Name
MutL protein homolog 1
Location in the cell
Cytoplasmic Reliability : 1.796 Nuclear Reliability : 1.77
Sequence
CDS
ATGAATGAGCCAGGAATAATTCGAAAACTGAGTGAAGAAGTTGTGAACCGTATTGCTGCTGGTGAAATAGTGCAAAGACCAGCTAATGCATTAAAAGAACTTATAGAAAACAGCTTGGATGCCAAATCAACTAACATCATTATTACAGTGAAGTCTGGTGGTTTGAAGTTCTTACAGATACAGGACAATGGGACAGGTATAAGAAATGAAGACTTAGATATCGTATGTGAGAGATTCACAACATCTAAACTCAGAAAATATGAGGACTTACAAGAAATCAGTACCTACGGATTTCGTGGAGAGGCTCTCGCGAGTATCAGTCATATTGCACACCTTACAATATTGACAAAAACTGCACAAGACAAATGTGCTTACAAAGCCTCATATGAAAATGGAAAACTGAAAGGCCCCATAAAAGCTTGTGCTGGCAACAACGGCACTCAGATAACCGTTGAAGATTTATTTTATAACGTTGTGGCACGTAAAGGGGCATTGCGTAGTCCTACAGAAGAATACACGCGTTTAATGGAAGTGGTCGGAGGATATGCTATACATAATAGTCAAGTTGGATTTACTTTGAAAAAATTCAGTGAGAATACTGACTTGAAAACAGCGATGAAATCATCTGTCATAGAAAATGTTAGAATTATTTATGGTAACACAATAGCAAGAGAGCTGATAGAGGTTGAAATTTTGGATGCAAACTTGAAGCTGTCGATACATGGATACATTACAAACGTTAATTATTCCAATAAGAAAGGGATACTGCTCCTGTTCATCAATCATAGGCTTGTGGATTCACAGGCAATAAGAAAAGCCGTGGATTCTGTTTACTCAACATATTTACCAAAAAACTCACACGCATTTGTATACTTGAGCATTGAATTAGATCCGAAAAACGTGGACGTCAACGTTCATCCAACGAAACACGAAGTTCAGTTTCTTTACGAGGAGCAAATCGTCGAGAAGATCAAGACGTGCATCGAAACGAAACTCTTGGGCTGTAGCTCCACGCGCGTGCTGTACACGCAGGCCCGCCTCCCCGGCGCCGCCCAGCCCAGGGACGTCGCGGACAAGCCCACGGACGCGGCAAAAACTTCCGCCGCACACATGGTACGCGTCAACCCTAACGTCCAGAAGATCGACAAGTTCTTTGAACTTGAACCAAATAAAACGAATCAACCGACTGGAGATAATGCGAGTGACGTAGTAAATGATGTAGATCAACAACAGAAAATACCTAGTGAACAAGACTTGAACAATTCTATAGTAAAAGAGCCAGCAGAAGATAATAAAGTACAAGAGAAATGGAAACAGTATACTGAGGCGGGCCAAGCCAACATTACGTACGTAGACCCTAAAGATTCCTTCAAGACTAGAACGTTTCAATACCAGAGGATAGAAACGAAACTGACCAGCGTAAAACAGCTCAGATTAGAAGTGGAGAACAATTGTAACTCAAATCTAAGGGAAATCTTGGCTAATTTAATATTCATAGCCTGTATAGATTGTCACAAGTCATTAATACAGCACTCTACGAAACTGTATCTTTGTGATACGACAAGATTGACGGAGGAATTGTTTTACGAGACATTACTATACGATTTTCAAAACTTCGGTTTCATCAAACTGTCGAATCCATTGCCATTGGAAGAATTATTTGTTCTTGGTCTGAACGGGAGCACTCAAAACTCCGAACAGGATTCTGAGCTCGGAGATACAGGAGAAATGGCTCAACATATGACCCAACTATTGTTGAGCAAACGCGCAATGCTCTACGAATACTTTTCTCTGGAAGTAAATGGAGAAGGAGAATTATTGGCCCTACCATTACTTTTGGATGGTCACGCTCCGTTCGTGGGTGCTTTACCGACGTATTTAGTCCGTCTCGTTACCGAAGTCAATTGGGATTCTGAAAAGGAGTGCTTTGAGACTTTCTGCAGACAGACGGCCAAGTTTTATGCTCAACCTAATCCGGACCCTAATGTCGAGAGCGTGTCGTCGGAGCACAGGAGACAAGAGCACGTGATATTCCCTGCTATAAGAAAATATTTCCTACCGCCAAGTAGCTTTATAAGTAATGGAGCTATACTGCAAATAGCTAGTTTAAATGATTTGTATAAGGTTTTTGAACGTTGTTGA
Protein
MNEPGIIRKLSEEVVNRIAAGEIVQRPANALKELIENSLDAKSTNIIITVKSGGLKFLQIQDNGTGIRNEDLDIVCERFTTSKLRKYEDLQEISTYGFRGEALASISHIAHLTILTKTAQDKCAYKASYENGKLKGPIKACAGNNGTQITVEDLFYNVVARKGALRSPTEEYTRLMEVVGGYAIHNSQVGFTLKKFSENTDLKTAMKSSVIENVRIIYGNTIARELIEVEILDANLKLSIHGYITNVNYSNKKGILLLFINHRLVDSQAIRKAVDSVYSTYLPKNSHAFVYLSIELDPKNVDVNVHPTKHEVQFLYEEQIVEKIKTCIETKLLGCSSTRVLYTQARLPGAAQPRDVADKPTDAAKTSAAHMVRVNPNVQKIDKFFELEPNKTNQPTGDNASDVVNDVDQQQKIPSEQDLNNSIVKEPAEDNKVQEKWKQYTEAGQANITYVDPKDSFKTRTFQYQRIETKLTSVKQLRLEVENNCNSNLREILANLIFIACIDCHKSLIQHSTKLYLCDTTRLTEELFYETLLYDFQNFGFIKLSNPLPLEELFVLGLNGSTQNSEQDSELGDTGEMAQHMTQLLLSKRAMLYEYFSLEVNGEGELLALPLLLDGHAPFVGALPTYLVRLVTEVNWDSEKECFETFCRQTAKFYAQPNPDPNVESVSSEHRRQEHVIFPAIRKYFLPPSSFISNGAILQIASLNDLYKVFERC
Summary
Description
Heterodimerizes with Pms2 to form MutL alpha, a component of the post-replicative DNA mismatch repair system (MMR). DNA repair is initiated by MutS alpha (Msh2-Msh6) or MutS beta (MSH2-MSH3) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of Pms2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (Mlh1-Pms2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in case of major DNA damages. Heterodimerizes with Mlh3 to form MutL gamma which plays a role in meiosis (By similarity).
Heterodimerizes with PMS2 to form MutL alpha, a component of the post-replicative DNA mismatch repair system (MMR). DNA repair is initiated by MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of PMS2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (MLH1-PMS2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in case of major DNA damages. Heterodimerizes with MLH3 to form MutL gamma which plays a role in meiosis.
Heterodimerizes with PMS2 to form MutL alpha, a component of the post-replicative DNA mismatch repair system (MMR). DNA repair is initiated by MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of PMS2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (MLH1-PMS2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in case of major DNA damages. Heterodimerizes with MLH3 to form MutL gamma which plays a role in meiosis.
Subunit
Component of the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1. Heterodimer of MLH1 and PMS2 (MutL alpha), MLH1 and PMS1 (MutL beta) or MLH1 and MLH3 (MutL gamma). Forms a ternary complex with MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3). Part of the BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the RAD50-MRE11-NBS1 protein complex. This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains. Interacts with MCM9; the interaction recruits MLH1 to chromatin. Interacts MCM8. Interacts with PMS2. Interacts with MBD4. Interacts with EXO1. Interacts with MTMR15/FAN1.
Component of the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1 (PubMed:26300262). Heterodimer of MLH1 and PMS2 (MutL alpha), MLH1 and PMS1 (MutL beta) or MLH1 and MLH3 (MutL gamma). Forms a ternary complex with MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3). Part of the BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the RAD50-MRE11-NBS1 protein complex (PubMed:10783165). This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains (PubMed:10097147). Interacts with MCM9; the interaction recruits MLH1 to chromatin (PubMed:26300262). Interacts MCM8 (PubMed:26300262). Interacts with PMS2 (PubMed:11427529, PubMed:22753075). Interacts with MBD4 (PubMed:10097147). Interacts with EXO1 (PubMed:11427529, PubMed:11429708, PubMed:12414623, PubMed:14676842, PubMed:22753075). Interacts with MTMR15/FAN1 (PubMed:20603073).
Component of the DNA mismatch repair (MMR) complex composed at least of MSH2, MSH3, MSH6, PMS1 and MLH1 (PubMed:26300262). Heterodimer of MLH1 and PMS2 (MutL alpha), MLH1 and PMS1 (MutL beta) or MLH1 and MLH3 (MutL gamma). Forms a ternary complex with MutS alpha (MSH2-MSH6) or MutS beta (MSH2-MSH3). Part of the BRCA1-associated genome surveillance complex (BASC), which contains BRCA1, MSH2, MSH6, MLH1, ATM, BLM, PMS2 and the RAD50-MRE11-NBS1 protein complex (PubMed:10783165). This association could be a dynamic process changing throughout the cell cycle and within subnuclear domains (PubMed:10097147). Interacts with MCM9; the interaction recruits MLH1 to chromatin (PubMed:26300262). Interacts MCM8 (PubMed:26300262). Interacts with PMS2 (PubMed:11427529, PubMed:22753075). Interacts with MBD4 (PubMed:10097147). Interacts with EXO1 (PubMed:11427529, PubMed:11429708, PubMed:12414623, PubMed:14676842, PubMed:22753075). Interacts with MTMR15/FAN1 (PubMed:20603073).
Similarity
Belongs to the DNA mismatch repair MutL/HexB family.
Keywords
ATP-binding
Cell cycle
Chromosome
Complete proteome
DNA damage
DNA repair
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
3D-structure
Acetylation
Alternative splicing
Disease mutation
Hereditary nonpolyposis colorectal cancer
Polymorphism
Tumor suppressor
Feature
chain DNA mismatch repair protein Mlh1
splice variant In isoform 2.
sequence variant In HNPCC2; dbSNP:rs367654552.
splice variant In isoform 2.
sequence variant In HNPCC2; dbSNP:rs367654552.
Uniprot
A0A194QIA4
A0A2A4J730
A0A2W1BJ81
A0A3S2M0G3
A0A2H1WVL2
A0A1A8MXV9
+ More
A0A1A8NKX3 A0A3Q1IF16 A0A1A8EP69 A0A1A8JBC5 A0A1A8CT25 A0A1A7ZFT8 A0A1A8D7L7 H2V8A2 A0A3B5BFC6 A0A3Q0R8T3 A0A3Q1ATQ8 A0A3Q3A5A3 A0A3P8UHI1 A0A147B2U5 I3JVJ6 A0A3Q3MJH2 A0A3P8WP48 A0A3Q2WPX6 A0A3P8QP15 A0A3P9BXD5 N6UIA2 A0A060VUS2 A0A1W4YPA0 A0A3B4CLB8 A0A2M4BGC7 A0A3Q1IEZ7 A0A1S3N2R6 A0A3Q2DRB8 A0A3P9MXL2 A0A195FX28 A0A3Q2VMU2 A0A3Q1FLH9 A0A3B4TLS6 A0A0F8C3Z0 A0A061I7W4 A0A3Q3IY45 A0A2I4AZJ2 A0A2U9CCK6 K7IN39 C3Y458 A0A3B4YQ70 A0A3B3XS72 A0A1W4YNV8 Q9JK91 A0A3Q3WPT4 V9KFJ8 A0A250Y541 A0A195CXK8 A0A087Y5T2 A0A1A7XZX0 A0A3B3CF03 I3M618 A0A061I5I4 A0A1S2ZC56 A0A3B3HRQ1 A0A2K6Q8L6 A0A2K5PMB6 A0A218UZY5 A0A3B3UVR8 A0A093GK37 A0A2K6EAM3 G7NYL5 A0A0D9RNU4 G7MJ24 A0A232F9W4 A0A151WW18 Q8VDI4 A0A2K5Y4B2 A0A2K5NFX5 A0A1U7THD7 A0A091FJK2 A0A2K5DJK9 A0A0M9ACB3 A0A2M4BGM0 I3LT92 A0A096MTT9 A0A2J8WYG5 A0A2K6TQ60 A0A154PF65 G1QYL9 U6CWA1 A0A2R9AM85 H9FUL2 H2QMA1 A0A3B4BG63 G3RJQ3 H0WYF6 A0A158P0T6 Q5GJ64 B2R6K0 P40692 W5MBU0 A0A2K5UB34 D3K5L8
A0A1A8NKX3 A0A3Q1IF16 A0A1A8EP69 A0A1A8JBC5 A0A1A8CT25 A0A1A7ZFT8 A0A1A8D7L7 H2V8A2 A0A3B5BFC6 A0A3Q0R8T3 A0A3Q1ATQ8 A0A3Q3A5A3 A0A3P8UHI1 A0A147B2U5 I3JVJ6 A0A3Q3MJH2 A0A3P8WP48 A0A3Q2WPX6 A0A3P8QP15 A0A3P9BXD5 N6UIA2 A0A060VUS2 A0A1W4YPA0 A0A3B4CLB8 A0A2M4BGC7 A0A3Q1IEZ7 A0A1S3N2R6 A0A3Q2DRB8 A0A3P9MXL2 A0A195FX28 A0A3Q2VMU2 A0A3Q1FLH9 A0A3B4TLS6 A0A0F8C3Z0 A0A061I7W4 A0A3Q3IY45 A0A2I4AZJ2 A0A2U9CCK6 K7IN39 C3Y458 A0A3B4YQ70 A0A3B3XS72 A0A1W4YNV8 Q9JK91 A0A3Q3WPT4 V9KFJ8 A0A250Y541 A0A195CXK8 A0A087Y5T2 A0A1A7XZX0 A0A3B3CF03 I3M618 A0A061I5I4 A0A1S2ZC56 A0A3B3HRQ1 A0A2K6Q8L6 A0A2K5PMB6 A0A218UZY5 A0A3B3UVR8 A0A093GK37 A0A2K6EAM3 G7NYL5 A0A0D9RNU4 G7MJ24 A0A232F9W4 A0A151WW18 Q8VDI4 A0A2K5Y4B2 A0A2K5NFX5 A0A1U7THD7 A0A091FJK2 A0A2K5DJK9 A0A0M9ACB3 A0A2M4BGM0 I3LT92 A0A096MTT9 A0A2J8WYG5 A0A2K6TQ60 A0A154PF65 G1QYL9 U6CWA1 A0A2R9AM85 H9FUL2 H2QMA1 A0A3B4BG63 G3RJQ3 H0WYF6 A0A158P0T6 Q5GJ64 B2R6K0 P40692 W5MBU0 A0A2K5UB34 D3K5L8
Pubmed
26354079
28756777
21551351
25186727
24487278
23537049
+ More
24755649 25835551 23929341 20075255 18563158 16141072 8674118 21183079 24402279 28087693 29451363 17554307 25362486 22002653 28648823 15489334 30723633 22722832 25319552 16136131 25463417 22398555 21347285 8145827 8128251 7812952 7757073 14702039 16641997 10783165 10097147 11427529 11429708 12414623 14676842 8751876 16873062 18206974 16188885 16873053 18157157 18669648 19690332 20603073 21269460 21406692 22223895 23186163 24275569 26300262 26249686 20020535 21120944 7661930 8571956 8566964 8872463 8797773 8609062 9311737 8993976 9087566 9218993 9048925 9272156 9067757 9259192 9298827 9399661 9032648 9326924 9718327 9611074 9559627 9526167 10627141 10660333 10671064 9833759 10375096 9927034 10573010 10323887 10480359 10598809 10386556 10413423 10777691 10713887 10882759 12132870 11369138 11389087 11726306 11555625 11139242 11748856 12095971 12373605 12115348 11781295 11839723 11793442 11754112 12200596 11870161 12362047 12658575 14504054 12655562 12655564 14635101 15139004 15365995 15365996 14961575 15064764 15184898 16083711 16451135 17440981 17510385 17301300 18033691 18561205 22753075 28494185
24755649 25835551 23929341 20075255 18563158 16141072 8674118 21183079 24402279 28087693 29451363 17554307 25362486 22002653 28648823 15489334 30723633 22722832 25319552 16136131 25463417 22398555 21347285 8145827 8128251 7812952 7757073 14702039 16641997 10783165 10097147 11427529 11429708 12414623 14676842 8751876 16873062 18206974 16188885 16873053 18157157 18669648 19690332 20603073 21269460 21406692 22223895 23186163 24275569 26300262 26249686 20020535 21120944 7661930 8571956 8566964 8872463 8797773 8609062 9311737 8993976 9087566 9218993 9048925 9272156 9067757 9259192 9298827 9399661 9032648 9326924 9718327 9611074 9559627 9526167 10627141 10660333 10671064 9833759 10375096 9927034 10573010 10323887 10480359 10598809 10386556 10413423 10777691 10713887 10882759 12132870 11369138 11389087 11726306 11555625 11139242 11748856 12095971 12373605 12115348 11781295 11839723 11793442 11754112 12200596 11870161 12362047 12658575 14504054 12655562 12655564 14635101 15139004 15365995 15365996 14961575 15064764 15184898 16083711 16451135 17440981 17510385 17301300 18033691 18561205 22753075 28494185
EMBL
KQ459193
KPJ03181.1
NWSH01002958
PCG67223.1
KZ150086
PZC73765.1
+ More
RSAL01000087 RVE48250.1 ODYU01011328 SOQ57006.1 HAEF01020185 SBR61344.1 HAEH01002739 SBR69534.1 HAEB01002140 SBQ48667.1 HAED01020495 SBR07066.1 HADZ01018004 SBP81945.1 HADY01003019 SBP41504.1 HAEA01001627 SBQ30107.1 GCES01001225 JAR85098.1 AERX01006539 AERX01006540 APGK01024678 KB740558 ENN80366.1 FR904307 CDQ58586.1 GGFJ01002951 MBW52092.1 KQ981200 KYN44993.1 KQ042804 KKF10442.1 KE674453 ERE76413.1 CP026257 AWP14307.1 GG666484 EEN65084.1 AF250844 AK168849 U60872 U59881 U59882 U59883 U59884 JW864554 AFO97071.1 GFFV01001291 JAV38654.1 KQ977141 KYN05381.1 AYCK01020172 HADX01001068 SBP23300.1 AGTP01006768 ERE76412.1 MUZQ01000082 OWK59218.1 KL215897 KFV67159.1 CM001277 EHH51433.1 AQIB01119299 CM001254 EHH16516.1 NNAY01000599 OXU27465.1 KQ982691 KYQ52073.1 BC021815 AAH21815.1 KL447035 KFO69289.1 KQ435706 KOX80229.1 GGFJ01003002 MBW52143.1 AEMK02000085 DQIR01169089 DQIR01215976 DQIR01230351 DQIR01246560 DQIR01268204 DQIR01268205 DQIR01268206 DQIR01289569 DQIR01292542 DQIR01293419 DQIR01298061 HDB24566.1 HDB85828.1 AHZZ02020311 NDHI03003375 PNJ74806.1 KQ434892 KZC10526.1 ADFV01172234 ADFV01172235 ADFV01172236 ADFV01172237 ADFV01172238 ADFV01172239 HAAF01002301 CCP74127.1 AJFE02091605 AJFE02091606 AJFE02091607 JU334568 JU475654 JV047104 AFE78321.1 AFH32458.1 AFI37175.1 AACZ04000223 AACZ04000224 GABC01008247 GABF01003843 GABD01008642 NBAG03000225 JAA03091.1 JAA18302.1 JAA24458.1 PNI73893.1 CABD030020751 CABD030020752 CABD030020753 CABD030020754 AAQR03005980 AAQR03005981 AAQR03005982 AAQR03005983 AAQR03005984 ADTU01005783 AY517558 AAT44531.1 AK312609 BAG35497.1 U07343 U07418 U40978 U40960 U40961 U40962 U40963 U40964 U40965 U40966 U40967 U40968 U40969 U40970 U40971 U40972 U40973 U40974 U40975 U40976 U40977 U17857 U17839 U17840 U17841 U17842 U17843 U17844 U17845 U17846 U17847 U17848 U17849 U17851 U17852 U17853 U17854 U17855 U17856 AY217549 AK295359 AK298324 AK298583 AK316074 AK316264 AC006583 AC011816 CH471055 BC006850 AHAT01021992 AQIA01039189 AQIA01039190 AQIA01039191 AQIA01039192 GU373696 ADC38896.1
RSAL01000087 RVE48250.1 ODYU01011328 SOQ57006.1 HAEF01020185 SBR61344.1 HAEH01002739 SBR69534.1 HAEB01002140 SBQ48667.1 HAED01020495 SBR07066.1 HADZ01018004 SBP81945.1 HADY01003019 SBP41504.1 HAEA01001627 SBQ30107.1 GCES01001225 JAR85098.1 AERX01006539 AERX01006540 APGK01024678 KB740558 ENN80366.1 FR904307 CDQ58586.1 GGFJ01002951 MBW52092.1 KQ981200 KYN44993.1 KQ042804 KKF10442.1 KE674453 ERE76413.1 CP026257 AWP14307.1 GG666484 EEN65084.1 AF250844 AK168849 U60872 U59881 U59882 U59883 U59884 JW864554 AFO97071.1 GFFV01001291 JAV38654.1 KQ977141 KYN05381.1 AYCK01020172 HADX01001068 SBP23300.1 AGTP01006768 ERE76412.1 MUZQ01000082 OWK59218.1 KL215897 KFV67159.1 CM001277 EHH51433.1 AQIB01119299 CM001254 EHH16516.1 NNAY01000599 OXU27465.1 KQ982691 KYQ52073.1 BC021815 AAH21815.1 KL447035 KFO69289.1 KQ435706 KOX80229.1 GGFJ01003002 MBW52143.1 AEMK02000085 DQIR01169089 DQIR01215976 DQIR01230351 DQIR01246560 DQIR01268204 DQIR01268205 DQIR01268206 DQIR01289569 DQIR01292542 DQIR01293419 DQIR01298061 HDB24566.1 HDB85828.1 AHZZ02020311 NDHI03003375 PNJ74806.1 KQ434892 KZC10526.1 ADFV01172234 ADFV01172235 ADFV01172236 ADFV01172237 ADFV01172238 ADFV01172239 HAAF01002301 CCP74127.1 AJFE02091605 AJFE02091606 AJFE02091607 JU334568 JU475654 JV047104 AFE78321.1 AFH32458.1 AFI37175.1 AACZ04000223 AACZ04000224 GABC01008247 GABF01003843 GABD01008642 NBAG03000225 JAA03091.1 JAA18302.1 JAA24458.1 PNI73893.1 CABD030020751 CABD030020752 CABD030020753 CABD030020754 AAQR03005980 AAQR03005981 AAQR03005982 AAQR03005983 AAQR03005984 ADTU01005783 AY517558 AAT44531.1 AK312609 BAG35497.1 U07343 U07418 U40978 U40960 U40961 U40962 U40963 U40964 U40965 U40966 U40967 U40968 U40969 U40970 U40971 U40972 U40973 U40974 U40975 U40976 U40977 U17857 U17839 U17840 U17841 U17842 U17843 U17844 U17845 U17846 U17847 U17848 U17849 U17851 U17852 U17853 U17854 U17855 U17856 AY217549 AK295359 AK298324 AK298583 AK316074 AK316264 AC006583 AC011816 CH471055 BC006850 AHAT01021992 AQIA01039189 AQIA01039190 AQIA01039191 AQIA01039192 GU373696 ADC38896.1
Proteomes
UP000053268
UP000218220
UP000283053
UP000265040
UP000005226
UP000261400
+ More
UP000261340 UP000257160 UP000264800 UP000265080 UP000005207 UP000261640 UP000265120 UP000264840 UP000265100 UP000265160 UP000019118 UP000193380 UP000192224 UP000261440 UP000087266 UP000265020 UP000242638 UP000078541 UP000257200 UP000261420 UP000030759 UP000261600 UP000192220 UP000246464 UP000002358 UP000001554 UP000261360 UP000261480 UP000000589 UP000261620 UP000078542 UP000028760 UP000261560 UP000005215 UP000079721 UP000001038 UP000233200 UP000233040 UP000197619 UP000261500 UP000053875 UP000233120 UP000009130 UP000029965 UP000215335 UP000075809 UP000233140 UP000233060 UP000189704 UP000053760 UP000233020 UP000053105 UP000008227 UP000028761 UP000233220 UP000076502 UP000001073 UP000240080 UP000002277 UP000261520 UP000001519 UP000005225 UP000005205 UP000005640 UP000018468 UP000233100
UP000261340 UP000257160 UP000264800 UP000265080 UP000005207 UP000261640 UP000265120 UP000264840 UP000265100 UP000265160 UP000019118 UP000193380 UP000192224 UP000261440 UP000087266 UP000265020 UP000242638 UP000078541 UP000257200 UP000261420 UP000030759 UP000261600 UP000192220 UP000246464 UP000002358 UP000001554 UP000261360 UP000261480 UP000000589 UP000261620 UP000078542 UP000028760 UP000261560 UP000005215 UP000079721 UP000001038 UP000233200 UP000233040 UP000197619 UP000261500 UP000053875 UP000233120 UP000009130 UP000029965 UP000215335 UP000075809 UP000233140 UP000233060 UP000189704 UP000053760 UP000233020 UP000053105 UP000008227 UP000028761 UP000233220 UP000076502 UP000001073 UP000240080 UP000002277 UP000261520 UP000001519 UP000005225 UP000005205 UP000005640 UP000018468 UP000233100
Interpro
Gene 3D
ProteinModelPortal
A0A194QIA4
A0A2A4J730
A0A2W1BJ81
A0A3S2M0G3
A0A2H1WVL2
A0A1A8MXV9
+ More
A0A1A8NKX3 A0A3Q1IF16 A0A1A8EP69 A0A1A8JBC5 A0A1A8CT25 A0A1A7ZFT8 A0A1A8D7L7 H2V8A2 A0A3B5BFC6 A0A3Q0R8T3 A0A3Q1ATQ8 A0A3Q3A5A3 A0A3P8UHI1 A0A147B2U5 I3JVJ6 A0A3Q3MJH2 A0A3P8WP48 A0A3Q2WPX6 A0A3P8QP15 A0A3P9BXD5 N6UIA2 A0A060VUS2 A0A1W4YPA0 A0A3B4CLB8 A0A2M4BGC7 A0A3Q1IEZ7 A0A1S3N2R6 A0A3Q2DRB8 A0A3P9MXL2 A0A195FX28 A0A3Q2VMU2 A0A3Q1FLH9 A0A3B4TLS6 A0A0F8C3Z0 A0A061I7W4 A0A3Q3IY45 A0A2I4AZJ2 A0A2U9CCK6 K7IN39 C3Y458 A0A3B4YQ70 A0A3B3XS72 A0A1W4YNV8 Q9JK91 A0A3Q3WPT4 V9KFJ8 A0A250Y541 A0A195CXK8 A0A087Y5T2 A0A1A7XZX0 A0A3B3CF03 I3M618 A0A061I5I4 A0A1S2ZC56 A0A3B3HRQ1 A0A2K6Q8L6 A0A2K5PMB6 A0A218UZY5 A0A3B3UVR8 A0A093GK37 A0A2K6EAM3 G7NYL5 A0A0D9RNU4 G7MJ24 A0A232F9W4 A0A151WW18 Q8VDI4 A0A2K5Y4B2 A0A2K5NFX5 A0A1U7THD7 A0A091FJK2 A0A2K5DJK9 A0A0M9ACB3 A0A2M4BGM0 I3LT92 A0A096MTT9 A0A2J8WYG5 A0A2K6TQ60 A0A154PF65 G1QYL9 U6CWA1 A0A2R9AM85 H9FUL2 H2QMA1 A0A3B4BG63 G3RJQ3 H0WYF6 A0A158P0T6 Q5GJ64 B2R6K0 P40692 W5MBU0 A0A2K5UB34 D3K5L8
A0A1A8NKX3 A0A3Q1IF16 A0A1A8EP69 A0A1A8JBC5 A0A1A8CT25 A0A1A7ZFT8 A0A1A8D7L7 H2V8A2 A0A3B5BFC6 A0A3Q0R8T3 A0A3Q1ATQ8 A0A3Q3A5A3 A0A3P8UHI1 A0A147B2U5 I3JVJ6 A0A3Q3MJH2 A0A3P8WP48 A0A3Q2WPX6 A0A3P8QP15 A0A3P9BXD5 N6UIA2 A0A060VUS2 A0A1W4YPA0 A0A3B4CLB8 A0A2M4BGC7 A0A3Q1IEZ7 A0A1S3N2R6 A0A3Q2DRB8 A0A3P9MXL2 A0A195FX28 A0A3Q2VMU2 A0A3Q1FLH9 A0A3B4TLS6 A0A0F8C3Z0 A0A061I7W4 A0A3Q3IY45 A0A2I4AZJ2 A0A2U9CCK6 K7IN39 C3Y458 A0A3B4YQ70 A0A3B3XS72 A0A1W4YNV8 Q9JK91 A0A3Q3WPT4 V9KFJ8 A0A250Y541 A0A195CXK8 A0A087Y5T2 A0A1A7XZX0 A0A3B3CF03 I3M618 A0A061I5I4 A0A1S2ZC56 A0A3B3HRQ1 A0A2K6Q8L6 A0A2K5PMB6 A0A218UZY5 A0A3B3UVR8 A0A093GK37 A0A2K6EAM3 G7NYL5 A0A0D9RNU4 G7MJ24 A0A232F9W4 A0A151WW18 Q8VDI4 A0A2K5Y4B2 A0A2K5NFX5 A0A1U7THD7 A0A091FJK2 A0A2K5DJK9 A0A0M9ACB3 A0A2M4BGM0 I3LT92 A0A096MTT9 A0A2J8WYG5 A0A2K6TQ60 A0A154PF65 G1QYL9 U6CWA1 A0A2R9AM85 H9FUL2 H2QMA1 A0A3B4BG63 G3RJQ3 H0WYF6 A0A158P0T6 Q5GJ64 B2R6K0 P40692 W5MBU0 A0A2K5UB34 D3K5L8
PDB
4P7A
E-value=2.91519e-129,
Score=1185
Ontologies
PATHWAY
GO
GO:0005524
GO:0032300
GO:0030983
GO:0016887
GO:0006298
GO:0016021
GO:0043060
GO:0007141
GO:0051026
GO:0007283
GO:0016321
GO:0032407
GO:0008630
GO:0045141
GO:0032137
GO:0000289
GO:0007129
GO:0032389
GO:0000795
GO:0016446
GO:0003697
GO:0001673
GO:0005654
GO:0048477
GO:0003682
GO:0006303
GO:0009617
GO:0048298
GO:0019899
GO:0045950
GO:0005712
GO:0051257
GO:0000712
GO:0045190
GO:0007060
GO:0005715
GO:0048304
GO:0007140
GO:0051321
GO:0005634
GO:0002204
GO:0007131
GO:0005694
GO:0016447
GO:0045143
GO:0006281
GO:0000794
GO:0000793
GO:0006974
GO:0045132
GO:0016020
GO:0003676
GO:0051287
GO:0030117
GO:0006418
GO:0003707
GO:0006259
PANTHER
Topology
Subcellular location
Nucleus
Recruited to chromatin in a MCM9-dependent manner. With evidence from 2 publications.
Chromosome Recruited to chromatin in a MCM9-dependent manner. With evidence from 2 publications.
Chromosome Recruited to chromatin in a MCM9-dependent manner. With evidence from 2 publications.
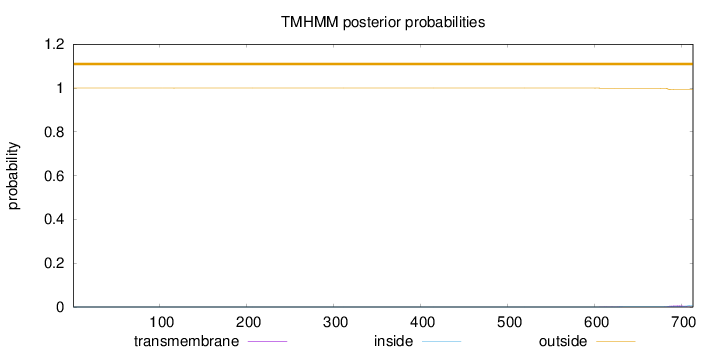
Length:
713
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15105
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00024
outside
1 - 713
Population Genetic Test Statistics
Pi
219.827901
Theta
179.197157
Tajima's D
0.75113
CLR
0.105628
CSRT
0.589720513974301
Interpretation
Uncertain
