Pre Gene Modal
BGIBMGA010687
Annotation
PREDICTED:_isocitrate_dehydrogenase_[NAD]_subunit_beta?_mitochondrial_[Bombyx_mori]
Full name
Isocitrate dehydrogenase [NAD] subunit, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.502 Mitochondrial Reliability : 1.787
Sequence
CDS
ATGTCCTTTCTTAGCAGGAATATTTTTCGAGCTGTTATGCAGGGATCTCAACATGTTGGAAAGGGTGTACACACAAGTTCGGTAACAACGGAGAAAAATCAACCGAGGGCAACAAAGGAAGGTCGTATCAAATGCACCTTGATTCCTGGGGACGGTGTAGGTCCTGAGCTTGTCTACGCTGTACAAGAAGTTTTCAAGGCGGCCAGCATTCCAGTGGACTTTGAATCCTTTTTCTTTTCTGAAGTGAATCCAACACTTAGTGCACCACTCGAAGACGTCGTAAACTCTATCGCTGTTAACAAGATTTGTATCAAGGGTATCTTAGCCACTCCTGACTTCTCTCACACTGGCGAGCTACAGACCCTAAACATGAAGCTCCGTAATGCCCTGGATCTTTACGCTAACGTGGTGCACGTGAAGTCATTGCCCAACGTGAAGTGCAGGCACCAGGACGTGGACTGCATCATCATCAGAGAACAGACCGAAGGAGAGTACTCAGCTTTGGAACATGAATCCGTTCCCGGCGTGGTGGAGTGTTTGAAGATCATCACCGCAGCGAAATCCGAGCGTATCGCGAAATTCGCTTTCGACTACGCCGTGAAGATGGGCCGCAAGAAGGTCACGGCTGTCCACAAGGCCAACATCATGAAGCTGGGCGACGGACTGTTCCTACGCAGCTGCGAGGAGATGGCCAAATTGTACCCTCGCATCCAGTTCGAGAAGATGATCGTGGACAACTGCACGATGCAGATGGTGTCCAACCCGAACCAGTTCGACGTGATGGTGACTCCGAACCTGTACGGGAACATCGTGGACAACCTGGCCAGCGGGCTGGTGGGCGGCGCCGGCGTGGTCGCGGGGGCCTCCTACAGCGCTGACTGCGCGGTCTTCGAACAGGGAGCAAGACACATCTTCTCCGGCGCCGTGGGCAAGAACATCGCGAACCCGACCGCCATGCTGCTCTGCTCCGCCAATCTCCTGGCGCACGTCAACCTGCCGCAGTACTCGAAGATGATCAAGAACGCCATCAATAAAGTTCTGAAAGACGCCAAAGTGCGAACAAAAGATCTTGGCGGTCAGTCCACCACCAAAGACTTCACAAACGCGGTCATTCACAGCCTCTCCTAA
Protein
MSFLSRNIFRAVMQGSQHVGKGVHTSSVTTEKNQPRATKEGRIKCTLIPGDGVGPELVYAVQEVFKAASIPVDFESFFFSEVNPTLSAPLEDVVNSIAVNKICIKGILATPDFSHTGELQTLNMKLRNALDLYANVVHVKSLPNVKCRHQDVDCIIIREQTEGEYSALEHESVPGVVECLKIITAAKSERIAKFAFDYAVKMGRKKVTAVHKANIMKLGDGLFLRSCEEMAKLYPRIQFEKMIVDNCTMQMVSNPNQFDVMVTPNLYGNIVDNLASGLVGGAGVVAGASYSADCAVFEQGARHIFSGAVGKNIANPTAMLLCSANLLAHVNLPQYSKMIKNAINKVLKDAKVRTKDLGGQSTTKDFTNAVIHSLS
Summary
Similarity
Belongs to the isocitrate and isopropylmalate dehydrogenases family.
Uniprot
A0A3S2NDG4
A0A194QCI2
H9JMD4
S4PY54
A0A2H1W5S1
A0A2A4K2Y8
+ More
A0A212EJ59 A0A2W1BDC2 A0A0N0PE34 D6X3C9 N6TM86 A0A0T6AU79 U4UM49 A0A2Z5REI6 A0A2J7QDT2 A0A067R3Z8 A0A1B6D7T9 Q29AP6 B4G640 T1PBM5 U4UNX5 A0A2A3E8P5 A0A088ABZ1 A0A1B6FXP9 A0A3B0KB74 A0A182JCX8 A0A0K8U7L4 A0A182PJP3 A0A0A1XFK3 A0A1W4VR89 U5EYC6 A0A084VI48 B4QZC5 B4NHQ8 B4HM75 A0A1Y1KU63 B4PKY9 A0A182JSC9 B3P8P4 Q9VD58 B4JRL0 A0A1B0CS47 T1DQ13 A0A182W7P8 A0A1L8E086 A0A182MZA9 A0A182QPR3 A0A2M3Z6S3 A0A182FNA2 W5JFN5 A0A182M7L5 A0A1Y9HFE0 A0A182XZN3 A0A2M4AJZ5 A0A2M4BRR8 W8BMQ8 A0A182UZ23 A0A182X1H6 A0A182L2J8 Q7Q3A3 A0A182TYG4 A0A182IEF7 A0A023ERZ7 F4WP27 B3LVK6 A0A0L0CR33 A0A0K8TMK2 A0A1S4FPD7 E2BB73 Q16TS5 A0A1I8PE26 A0A195E662 A0A151WM03 A0A154PGB9 A0A0L7RC17 A0A0J7KKC3 A0A195EU14 A0A336K9V2 B4M0N8 A0A0L7KXY3 B4KAP7 A0A0N0BDP8 A0A0C9QMZ8 E2B128 E9IQS1 A0A1J1HY37 A0A232F7A0 A0A195B944 A0A1S3DA33 A0A0M4F6N4 A0A1Q3FYP7 A0A195CN40 K7IP35 A0A224XRZ2 T1HLD4 A0A0P4VMJ1 A0A1A9VAG9 A0A1A9Y0Z7 A0A1B0BMJ9 Q0QHL0
A0A212EJ59 A0A2W1BDC2 A0A0N0PE34 D6X3C9 N6TM86 A0A0T6AU79 U4UM49 A0A2Z5REI6 A0A2J7QDT2 A0A067R3Z8 A0A1B6D7T9 Q29AP6 B4G640 T1PBM5 U4UNX5 A0A2A3E8P5 A0A088ABZ1 A0A1B6FXP9 A0A3B0KB74 A0A182JCX8 A0A0K8U7L4 A0A182PJP3 A0A0A1XFK3 A0A1W4VR89 U5EYC6 A0A084VI48 B4QZC5 B4NHQ8 B4HM75 A0A1Y1KU63 B4PKY9 A0A182JSC9 B3P8P4 Q9VD58 B4JRL0 A0A1B0CS47 T1DQ13 A0A182W7P8 A0A1L8E086 A0A182MZA9 A0A182QPR3 A0A2M3Z6S3 A0A182FNA2 W5JFN5 A0A182M7L5 A0A1Y9HFE0 A0A182XZN3 A0A2M4AJZ5 A0A2M4BRR8 W8BMQ8 A0A182UZ23 A0A182X1H6 A0A182L2J8 Q7Q3A3 A0A182TYG4 A0A182IEF7 A0A023ERZ7 F4WP27 B3LVK6 A0A0L0CR33 A0A0K8TMK2 A0A1S4FPD7 E2BB73 Q16TS5 A0A1I8PE26 A0A195E662 A0A151WM03 A0A154PGB9 A0A0L7RC17 A0A0J7KKC3 A0A195EU14 A0A336K9V2 B4M0N8 A0A0L7KXY3 B4KAP7 A0A0N0BDP8 A0A0C9QMZ8 E2B128 E9IQS1 A0A1J1HY37 A0A232F7A0 A0A195B944 A0A1S3DA33 A0A0M4F6N4 A0A1Q3FYP7 A0A195CN40 K7IP35 A0A224XRZ2 T1HLD4 A0A0P4VMJ1 A0A1A9VAG9 A0A1A9Y0Z7 A0A1B0BMJ9 Q0QHL0
Pubmed
26354079
19121390
23622113
22118469
28756777
18362917
+ More
19820115 23537049 26760975 24845553 15632085 17994087 25315136 25830018 24438588 28004739 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25244985 24495485 20966253 12364791 14747013 17210077 24945155 21719571 26108605 26369729 20798317 17510324 18057021 26227816 21282665 28648823 20075255 27129103 20353571
19820115 23537049 26760975 24845553 15632085 17994087 25315136 25830018 24438588 28004739 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25244985 24495485 20966253 12364791 14747013 17210077 24945155 21719571 26108605 26369729 20798317 17510324 18057021 26227816 21282665 28648823 20075255 27129103 20353571
EMBL
RSAL01000087
RVE48253.1
KQ459193
KPJ03177.1
BABH01040060
BABH01040061
+ More
BABH01040062 GAIX01003883 JAA88677.1 ODYU01006505 SOQ48408.1 NWSH01000220 PCG78264.1 AGBW02014523 OWR41515.1 KZ150431 PZC70900.1 KQ459941 KPJ19165.1 KQ971372 EFA10361.1 APGK01058976 KB741292 ENN70365.1 LJIG01022860 KRT78350.1 KB632383 ERL94192.1 FX983738 BAX07237.1 NEVH01015814 PNF26733.1 KK852714 KDR17889.1 GEDC01015582 JAS21716.1 CM000070 EAL27303.2 CH479179 EDW23805.1 KA645510 AFP60139.1 ERL94193.1 KZ288338 PBC27834.1 GECZ01014802 JAS54967.1 OUUW01000007 SPP82926.1 GDHF01029655 JAI22659.1 GBXI01004576 JAD09716.1 GANO01002083 JAB57788.1 ATLV01013283 KE524849 KFB37642.1 CM000364 EDX13861.1 CH964272 EDW84668.1 CH480815 EDW43123.1 GEZM01075759 JAV63971.1 CM000160 EDW97938.1 CH954182 EDV54208.1 AE014297 BT004877 AAF55942.1 AAO45233.1 ACZ94978.1 CH916373 EDV94400.1 AJWK01025642 GAMD01002849 JAA98741.1 GFDF01001973 JAV12111.1 AXCN02002070 GGFM01003454 MBW24205.1 ADMH02001533 ETN62153.1 AXCM01004662 GGFK01007789 MBW41110.1 GGFJ01006532 MBW55673.1 GAMC01004170 JAC02386.1 AAAB01008964 EAA12327.4 APCN01005201 GAPW01002108 GAPW01002107 JAC11491.1 GL888243 EGI63958.1 CH902617 EDV42576.1 JRES01000142 KNC33874.1 GDAI01002019 JAI15584.1 GL446946 EFN87046.1 CH477641 EAT37918.1 KQ979608 KYN20367.1 KQ982944 KYQ48909.1 KQ434899 KZC10926.1 KQ414617 KOC68306.1 LBMM01006354 KMQ90674.1 KQ981965 KYN31740.1 UFQS01000198 UFQT01000198 SSX01217.1 SSX21597.1 CH940650 EDW67330.1 KRF83249.1 JTDY01004670 KOB67894.1 CH933806 EDW16784.1 KQ435851 KOX70768.1 GBYB01004929 JAG74696.1 GL444771 EFN60601.1 GL764900 EFZ17083.1 CVRI01000021 CRK91446.1 NNAY01000792 OXU26472.1 KQ976542 KYM81056.1 CP012526 ALC47395.1 GFDL01002395 JAV32650.1 KQ977574 KYN01887.1 GFTR01005677 JAW10749.1 ACPB03010980 GDKW01002551 JAI54044.1 JXJN01016973 CCAG010008224 DQ374009 EZ422365 ABG77987.1 ADD18641.1
BABH01040062 GAIX01003883 JAA88677.1 ODYU01006505 SOQ48408.1 NWSH01000220 PCG78264.1 AGBW02014523 OWR41515.1 KZ150431 PZC70900.1 KQ459941 KPJ19165.1 KQ971372 EFA10361.1 APGK01058976 KB741292 ENN70365.1 LJIG01022860 KRT78350.1 KB632383 ERL94192.1 FX983738 BAX07237.1 NEVH01015814 PNF26733.1 KK852714 KDR17889.1 GEDC01015582 JAS21716.1 CM000070 EAL27303.2 CH479179 EDW23805.1 KA645510 AFP60139.1 ERL94193.1 KZ288338 PBC27834.1 GECZ01014802 JAS54967.1 OUUW01000007 SPP82926.1 GDHF01029655 JAI22659.1 GBXI01004576 JAD09716.1 GANO01002083 JAB57788.1 ATLV01013283 KE524849 KFB37642.1 CM000364 EDX13861.1 CH964272 EDW84668.1 CH480815 EDW43123.1 GEZM01075759 JAV63971.1 CM000160 EDW97938.1 CH954182 EDV54208.1 AE014297 BT004877 AAF55942.1 AAO45233.1 ACZ94978.1 CH916373 EDV94400.1 AJWK01025642 GAMD01002849 JAA98741.1 GFDF01001973 JAV12111.1 AXCN02002070 GGFM01003454 MBW24205.1 ADMH02001533 ETN62153.1 AXCM01004662 GGFK01007789 MBW41110.1 GGFJ01006532 MBW55673.1 GAMC01004170 JAC02386.1 AAAB01008964 EAA12327.4 APCN01005201 GAPW01002108 GAPW01002107 JAC11491.1 GL888243 EGI63958.1 CH902617 EDV42576.1 JRES01000142 KNC33874.1 GDAI01002019 JAI15584.1 GL446946 EFN87046.1 CH477641 EAT37918.1 KQ979608 KYN20367.1 KQ982944 KYQ48909.1 KQ434899 KZC10926.1 KQ414617 KOC68306.1 LBMM01006354 KMQ90674.1 KQ981965 KYN31740.1 UFQS01000198 UFQT01000198 SSX01217.1 SSX21597.1 CH940650 EDW67330.1 KRF83249.1 JTDY01004670 KOB67894.1 CH933806 EDW16784.1 KQ435851 KOX70768.1 GBYB01004929 JAG74696.1 GL444771 EFN60601.1 GL764900 EFZ17083.1 CVRI01000021 CRK91446.1 NNAY01000792 OXU26472.1 KQ976542 KYM81056.1 CP012526 ALC47395.1 GFDL01002395 JAV32650.1 KQ977574 KYN01887.1 GFTR01005677 JAW10749.1 ACPB03010980 GDKW01002551 JAI54044.1 JXJN01016973 CCAG010008224 DQ374009 EZ422365 ABG77987.1 ADD18641.1
Proteomes
UP000283053
UP000053268
UP000005204
UP000218220
UP000007151
UP000053240
+ More
UP000007266 UP000019118 UP000030742 UP000235965 UP000027135 UP000001819 UP000008744 UP000095301 UP000242457 UP000005203 UP000268350 UP000075880 UP000075885 UP000192221 UP000030765 UP000000304 UP000007798 UP000001292 UP000002282 UP000075881 UP000008711 UP000000803 UP000001070 UP000092461 UP000075920 UP000075884 UP000075886 UP000069272 UP000000673 UP000075883 UP000075900 UP000076408 UP000075903 UP000076407 UP000075882 UP000007062 UP000075902 UP000075840 UP000007755 UP000007801 UP000037069 UP000008237 UP000008820 UP000095300 UP000078492 UP000075809 UP000076502 UP000053825 UP000036403 UP000078541 UP000008792 UP000037510 UP000009192 UP000053105 UP000000311 UP000183832 UP000215335 UP000078540 UP000079169 UP000092553 UP000078542 UP000002358 UP000015103 UP000078200 UP000092443 UP000092460 UP000092444
UP000007266 UP000019118 UP000030742 UP000235965 UP000027135 UP000001819 UP000008744 UP000095301 UP000242457 UP000005203 UP000268350 UP000075880 UP000075885 UP000192221 UP000030765 UP000000304 UP000007798 UP000001292 UP000002282 UP000075881 UP000008711 UP000000803 UP000001070 UP000092461 UP000075920 UP000075884 UP000075886 UP000069272 UP000000673 UP000075883 UP000075900 UP000076408 UP000075903 UP000076407 UP000075882 UP000007062 UP000075902 UP000075840 UP000007755 UP000007801 UP000037069 UP000008237 UP000008820 UP000095300 UP000078492 UP000075809 UP000076502 UP000053825 UP000036403 UP000078541 UP000008792 UP000037510 UP000009192 UP000053105 UP000000311 UP000183832 UP000215335 UP000078540 UP000079169 UP000092553 UP000078542 UP000002358 UP000015103 UP000078200 UP000092443 UP000092460 UP000092444
PRIDE
Pfam
PF00180 Iso_dh
Interpro
ProteinModelPortal
A0A3S2NDG4
A0A194QCI2
H9JMD4
S4PY54
A0A2H1W5S1
A0A2A4K2Y8
+ More
A0A212EJ59 A0A2W1BDC2 A0A0N0PE34 D6X3C9 N6TM86 A0A0T6AU79 U4UM49 A0A2Z5REI6 A0A2J7QDT2 A0A067R3Z8 A0A1B6D7T9 Q29AP6 B4G640 T1PBM5 U4UNX5 A0A2A3E8P5 A0A088ABZ1 A0A1B6FXP9 A0A3B0KB74 A0A182JCX8 A0A0K8U7L4 A0A182PJP3 A0A0A1XFK3 A0A1W4VR89 U5EYC6 A0A084VI48 B4QZC5 B4NHQ8 B4HM75 A0A1Y1KU63 B4PKY9 A0A182JSC9 B3P8P4 Q9VD58 B4JRL0 A0A1B0CS47 T1DQ13 A0A182W7P8 A0A1L8E086 A0A182MZA9 A0A182QPR3 A0A2M3Z6S3 A0A182FNA2 W5JFN5 A0A182M7L5 A0A1Y9HFE0 A0A182XZN3 A0A2M4AJZ5 A0A2M4BRR8 W8BMQ8 A0A182UZ23 A0A182X1H6 A0A182L2J8 Q7Q3A3 A0A182TYG4 A0A182IEF7 A0A023ERZ7 F4WP27 B3LVK6 A0A0L0CR33 A0A0K8TMK2 A0A1S4FPD7 E2BB73 Q16TS5 A0A1I8PE26 A0A195E662 A0A151WM03 A0A154PGB9 A0A0L7RC17 A0A0J7KKC3 A0A195EU14 A0A336K9V2 B4M0N8 A0A0L7KXY3 B4KAP7 A0A0N0BDP8 A0A0C9QMZ8 E2B128 E9IQS1 A0A1J1HY37 A0A232F7A0 A0A195B944 A0A1S3DA33 A0A0M4F6N4 A0A1Q3FYP7 A0A195CN40 K7IP35 A0A224XRZ2 T1HLD4 A0A0P4VMJ1 A0A1A9VAG9 A0A1A9Y0Z7 A0A1B0BMJ9 Q0QHL0
A0A212EJ59 A0A2W1BDC2 A0A0N0PE34 D6X3C9 N6TM86 A0A0T6AU79 U4UM49 A0A2Z5REI6 A0A2J7QDT2 A0A067R3Z8 A0A1B6D7T9 Q29AP6 B4G640 T1PBM5 U4UNX5 A0A2A3E8P5 A0A088ABZ1 A0A1B6FXP9 A0A3B0KB74 A0A182JCX8 A0A0K8U7L4 A0A182PJP3 A0A0A1XFK3 A0A1W4VR89 U5EYC6 A0A084VI48 B4QZC5 B4NHQ8 B4HM75 A0A1Y1KU63 B4PKY9 A0A182JSC9 B3P8P4 Q9VD58 B4JRL0 A0A1B0CS47 T1DQ13 A0A182W7P8 A0A1L8E086 A0A182MZA9 A0A182QPR3 A0A2M3Z6S3 A0A182FNA2 W5JFN5 A0A182M7L5 A0A1Y9HFE0 A0A182XZN3 A0A2M4AJZ5 A0A2M4BRR8 W8BMQ8 A0A182UZ23 A0A182X1H6 A0A182L2J8 Q7Q3A3 A0A182TYG4 A0A182IEF7 A0A023ERZ7 F4WP27 B3LVK6 A0A0L0CR33 A0A0K8TMK2 A0A1S4FPD7 E2BB73 Q16TS5 A0A1I8PE26 A0A195E662 A0A151WM03 A0A154PGB9 A0A0L7RC17 A0A0J7KKC3 A0A195EU14 A0A336K9V2 B4M0N8 A0A0L7KXY3 B4KAP7 A0A0N0BDP8 A0A0C9QMZ8 E2B128 E9IQS1 A0A1J1HY37 A0A232F7A0 A0A195B944 A0A1S3DA33 A0A0M4F6N4 A0A1Q3FYP7 A0A195CN40 K7IP35 A0A224XRZ2 T1HLD4 A0A0P4VMJ1 A0A1A9VAG9 A0A1A9Y0Z7 A0A1B0BMJ9 Q0QHL0
PDB
5YVT
E-value=5.15455e-85,
Score=801
Ontologies
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Mitochondrion
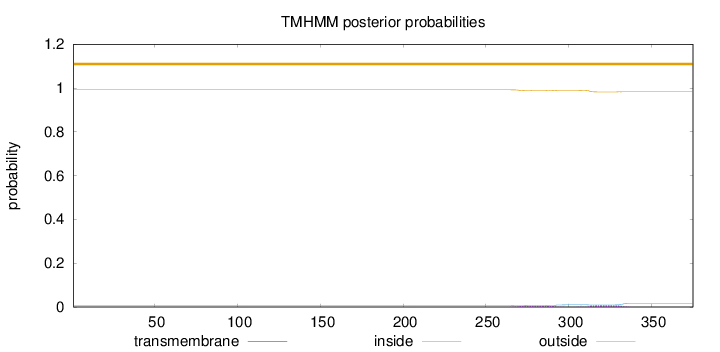
Length:
375
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29235
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00799
outside
1 - 375
Population Genetic Test Statistics
Pi
258.679459
Theta
165.437778
Tajima's D
1.126893
CLR
0.265352
CSRT
0.694765261736913
Interpretation
Uncertain
