Gene
KWMTBOMO15649
Pre Gene Modal
BGIBMGA010698
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_solute_carrier_family_10_member_6-like_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.11
Sequence
CDS
ATGGAAGAATACAAATACGTCGATGTATCTATTGAAGGTTCGGGCTTGGAGTTAAATGACGCGTTCAAAATCAGTTCGCAAAATGAGCAACTGGCCTGGGCCGAATGGAACTCGTCGAACCTTTACGAAAAACACGACAGCACGTGGAAGGGCAAGATACGCGTTAATGGAAATTTTATAGGTAGGACAAACTTGGTACTGGAAGTAAAAAGGGACGCCGATACGTTTTCCGTCACCAACGGTACGGTGCCCGTGGTGATCACGAGGGTCGAACGGGTCATAGACACGATATTCACCACCAGCGTTGCTGTGTTCATAGCTTTAATCTTTATCAATTTCGGATGTGCGATGCACTGGCCGACGGTGCGTGACGTCATCAAGCGACCGATCGGACCCGCCATAGGAATGGTCGGACAATTCCTTTTTATGCCACTCAGTTCCTTCGGTCTGGGCTACGTGATATTCCCGAACTCTCCCGCGATGCGTCTCGGGATGTTCTTCACGGGCGTAGCCCCCGGGGGCGGCGCCTCCAACGTCTGGACCTTCATCCTCGGCGGTAACCTCAACCTGTCCCTCGCCATGACGACCATATCTACTATTTGCTCATTTGGTAAGTGTTATTAA
Protein
MEEYKYVDVSIEGSGLELNDAFKISSQNEQLAWAEWNSSNLYEKHDSTWKGKIRVNGNFIGRTNLVLEVKRDADTFSVTNGTVPVVITRVERVIDTIFTTSVAVFIALIFINFGCAMHWPTVRDVIKRPIGPAIGMVGQFLFMPLSSFGLGYVIFPNSPAMRLGMFFTGVAPGGGASNVWTFILGGNLNLSLAMTTISTICSFGKCY
Summary
Uniprot
A0A2A4K2W2
A0A2A4K2I8
A0A2W1BC49
A0A2H1W5N5
A0A194QE01
A0A0N1IAA5
+ More
A0A3S2M0E9 A0A1S4FW99 Q16M32 A0A182GHY1 A0A023ESG9 A0A1Y1NGL9 A0A1Y1NM71 A0A1Y1NGD6 A0A1Y1NHG5 A0A1Y1NJA7 A0A1Y1NGD0 A0A1Q3FJB8 A0A1Q3FJ43 A0A182JTU0 A0A182IJS5 A0A182VZY7 A0A0C9RUD5 A0A084W717 B0W6D5 A0A182P3V4 E2AUA1 A0A182Y6Q6 A0A182M1T4 A0A182UNQ8 Q5TTX4 A0A0J7KDR9 A0A182HSC6 A0A182XAI6 A0A182FAH2 T1E8U9 A0A182U9H6 A0A182LEB8 W5JAK3 A0A182QJW2 A0A2M3Z129 U5ESB4 A0A2M4BMH5 A0A2M4BM28 A0A2M4BMM0 A0A182NR99 A0A158P2A0 A0A195E4D4 A0A182RQL2 E2C029 A0A1L8DE01 A0A1J1IXH4 A0A2M4AD31 A0A2M4ACT4 A0A1L8DEC7 A0A1L8DDY3 A0A1L8DE06 A0A1B0GKB1 A0A1B0EWL2 A0A067RI22 W8BLL6 A0A195FIT7 A0A2M4ABC1 V5GU37 A0A3Q0J802 F4W6J5 A0A1I8MQ07 A0A1B6CY20 A0A2M4ABD5 D6WZN2 A0A026WF95 A0A1I8NKQ8 A0A195BT61 A0A2J7PTH0 A0A2J7PTH4 K7J059 A0A195D319 A0A232FCH1 A0A1B0DC13 A0A1I8Q5L4 A0A1I8NEP0 A0A1W4WPR9 B4M6U9 A0A146LKT8 A0A0K8SEF7 A0A0A9YHI4 A0A1A9UM78 A0A034W221 A0A0K8U568 A0A336MLN3 A0A1A9WDB0 A0A336M0W0 D3TPB7 E0VCS0 A0A1A9Z8L0 A0A1B6F323 A0A0A1XB07 A0A224XJS0
A0A3S2M0E9 A0A1S4FW99 Q16M32 A0A182GHY1 A0A023ESG9 A0A1Y1NGL9 A0A1Y1NM71 A0A1Y1NGD6 A0A1Y1NHG5 A0A1Y1NJA7 A0A1Y1NGD0 A0A1Q3FJB8 A0A1Q3FJ43 A0A182JTU0 A0A182IJS5 A0A182VZY7 A0A0C9RUD5 A0A084W717 B0W6D5 A0A182P3V4 E2AUA1 A0A182Y6Q6 A0A182M1T4 A0A182UNQ8 Q5TTX4 A0A0J7KDR9 A0A182HSC6 A0A182XAI6 A0A182FAH2 T1E8U9 A0A182U9H6 A0A182LEB8 W5JAK3 A0A182QJW2 A0A2M3Z129 U5ESB4 A0A2M4BMH5 A0A2M4BM28 A0A2M4BMM0 A0A182NR99 A0A158P2A0 A0A195E4D4 A0A182RQL2 E2C029 A0A1L8DE01 A0A1J1IXH4 A0A2M4AD31 A0A2M4ACT4 A0A1L8DEC7 A0A1L8DDY3 A0A1L8DE06 A0A1B0GKB1 A0A1B0EWL2 A0A067RI22 W8BLL6 A0A195FIT7 A0A2M4ABC1 V5GU37 A0A3Q0J802 F4W6J5 A0A1I8MQ07 A0A1B6CY20 A0A2M4ABD5 D6WZN2 A0A026WF95 A0A1I8NKQ8 A0A195BT61 A0A2J7PTH0 A0A2J7PTH4 K7J059 A0A195D319 A0A232FCH1 A0A1B0DC13 A0A1I8Q5L4 A0A1I8NEP0 A0A1W4WPR9 B4M6U9 A0A146LKT8 A0A0K8SEF7 A0A0A9YHI4 A0A1A9UM78 A0A034W221 A0A0K8U568 A0A336MLN3 A0A1A9WDB0 A0A336M0W0 D3TPB7 E0VCS0 A0A1A9Z8L0 A0A1B6F323 A0A0A1XB07 A0A224XJS0
Pubmed
28756777
26354079
17510324
26483478
24945155
28004739
+ More
24438588 20798317 25244985 12364791 14747013 17210077 20966253 20920257 23761445 21347285 24845553 24495485 21719571 25315136 18362917 19820115 24508170 30249741 20075255 28648823 17994087 26823975 25401762 25348373 20353571 20566863 25830018
24438588 20798317 25244985 12364791 14747013 17210077 20966253 20920257 23761445 21347285 24845553 24495485 21719571 25315136 18362917 19820115 24508170 30249741 20075255 28648823 17994087 26823975 25401762 25348373 20353571 20566863 25830018
EMBL
NWSH01000220
PCG78268.1
PCG78269.1
KZ150431
PZC70897.1
ODYU01006505
+ More
SOQ48399.1 KQ459193 KPJ03175.1 KQ459941 KPJ19164.1 RSAL01000087 RVE48255.1 CH477882 EAT35385.1 JXUM01064723 KQ562312 KXJ76176.1 GAPW01001406 JAC12192.1 GEZM01003016 GEZM01003014 JAV96981.1 GEZM01003018 JAV96977.1 GEZM01003013 JAV96982.1 GEZM01003017 JAV96978.1 GEZM01003015 JAV96980.1 GEZM01003012 JAV96983.1 GFDL01007469 JAV27576.1 GFDL01007456 JAV27589.1 GBYB01012430 GBYB01012432 JAG82197.1 JAG82199.1 ATLV01021094 KE525312 KFB46011.1 DS231848 EDS36609.1 GL442815 EFN62958.1 AXCM01008428 AAAB01008859 EAL40865.4 LBMM01008848 KMQ88588.1 APCN01000261 GAMD01001877 JAA99713.1 ADMH02002037 ETN59814.1 AXCN02001467 GGFM01001478 MBW22229.1 GANO01003319 JAB56552.1 GGFJ01005053 MBW54194.1 GGFJ01004999 MBW54140.1 GGFJ01005131 MBW54272.1 ADTU01007043 KQ979657 KYN19941.1 GL451712 EFN78725.1 GFDF01009403 JAV04681.1 CVRI01000063 CRL04901.1 GGFK01005311 MBW38632.1 GGFK01005276 MBW38597.1 GFDF01009400 JAV04684.1 GFDF01009401 JAV04683.1 GFDF01009402 JAV04682.1 AJWK01028644 AJWK01028643 KK852620 KDR20050.1 GAMC01012300 GAMC01012299 JAB94256.1 KQ981523 KYN40313.1 GGFK01004691 MBW38012.1 GALX01000792 JAB67674.1 GL887707 EGI70330.1 GEDC01018929 JAS18369.1 GGFK01004748 MBW38069.1 KQ971372 EFA09677.1 KK107250 QOIP01000002 EZA54356.1 RLU25484.1 KQ976408 KYM91126.1 NEVH01021572 PNF19630.1 PNF19631.1 KQ976885 KYN07292.1 NNAY01000467 OXU28173.1 AJVK01030485 CH940653 EDW62516.1 GDHC01013476 GDHC01010051 JAQ05153.1 JAQ08578.1 GBRD01014150 JAG51676.1 GBHO01013051 GBHO01013041 GBHO01013037 GBHO01013030 GBRD01014151 JAG30553.1 JAG30563.1 JAG30567.1 JAG30574.1 JAG51675.1 GAKP01011139 GAKP01011138 JAC47813.1 GDHF01030520 GDHF01030097 GDHF01028155 JAI21794.1 JAI22217.1 JAI24159.1 UFQT01001481 SSX30735.1 UFQT01000396 SSX23966.1 CCAG010018936 EZ423269 ADD19545.1 DS235063 EEB11176.1 GECZ01025194 JAS44575.1 GBXI01005788 JAD08504.1 GFTR01006358 JAW10068.1
SOQ48399.1 KQ459193 KPJ03175.1 KQ459941 KPJ19164.1 RSAL01000087 RVE48255.1 CH477882 EAT35385.1 JXUM01064723 KQ562312 KXJ76176.1 GAPW01001406 JAC12192.1 GEZM01003016 GEZM01003014 JAV96981.1 GEZM01003018 JAV96977.1 GEZM01003013 JAV96982.1 GEZM01003017 JAV96978.1 GEZM01003015 JAV96980.1 GEZM01003012 JAV96983.1 GFDL01007469 JAV27576.1 GFDL01007456 JAV27589.1 GBYB01012430 GBYB01012432 JAG82197.1 JAG82199.1 ATLV01021094 KE525312 KFB46011.1 DS231848 EDS36609.1 GL442815 EFN62958.1 AXCM01008428 AAAB01008859 EAL40865.4 LBMM01008848 KMQ88588.1 APCN01000261 GAMD01001877 JAA99713.1 ADMH02002037 ETN59814.1 AXCN02001467 GGFM01001478 MBW22229.1 GANO01003319 JAB56552.1 GGFJ01005053 MBW54194.1 GGFJ01004999 MBW54140.1 GGFJ01005131 MBW54272.1 ADTU01007043 KQ979657 KYN19941.1 GL451712 EFN78725.1 GFDF01009403 JAV04681.1 CVRI01000063 CRL04901.1 GGFK01005311 MBW38632.1 GGFK01005276 MBW38597.1 GFDF01009400 JAV04684.1 GFDF01009401 JAV04683.1 GFDF01009402 JAV04682.1 AJWK01028644 AJWK01028643 KK852620 KDR20050.1 GAMC01012300 GAMC01012299 JAB94256.1 KQ981523 KYN40313.1 GGFK01004691 MBW38012.1 GALX01000792 JAB67674.1 GL887707 EGI70330.1 GEDC01018929 JAS18369.1 GGFK01004748 MBW38069.1 KQ971372 EFA09677.1 KK107250 QOIP01000002 EZA54356.1 RLU25484.1 KQ976408 KYM91126.1 NEVH01021572 PNF19630.1 PNF19631.1 KQ976885 KYN07292.1 NNAY01000467 OXU28173.1 AJVK01030485 CH940653 EDW62516.1 GDHC01013476 GDHC01010051 JAQ05153.1 JAQ08578.1 GBRD01014150 JAG51676.1 GBHO01013051 GBHO01013041 GBHO01013037 GBHO01013030 GBRD01014151 JAG30553.1 JAG30563.1 JAG30567.1 JAG30574.1 JAG51675.1 GAKP01011139 GAKP01011138 JAC47813.1 GDHF01030520 GDHF01030097 GDHF01028155 JAI21794.1 JAI22217.1 JAI24159.1 UFQT01001481 SSX30735.1 UFQT01000396 SSX23966.1 CCAG010018936 EZ423269 ADD19545.1 DS235063 EEB11176.1 GECZ01025194 JAS44575.1 GBXI01005788 JAD08504.1 GFTR01006358 JAW10068.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000008820
UP000069940
+ More
UP000249989 UP000075881 UP000075880 UP000075920 UP000030765 UP000002320 UP000075885 UP000000311 UP000076408 UP000075883 UP000075903 UP000007062 UP000036403 UP000075840 UP000076407 UP000069272 UP000075902 UP000075882 UP000000673 UP000075886 UP000075884 UP000005205 UP000078492 UP000075900 UP000008237 UP000183832 UP000092461 UP000027135 UP000078541 UP000079169 UP000007755 UP000095301 UP000007266 UP000053097 UP000279307 UP000078540 UP000235965 UP000002358 UP000078542 UP000215335 UP000092462 UP000095300 UP000192223 UP000008792 UP000078200 UP000091820 UP000092444 UP000009046 UP000092445
UP000249989 UP000075881 UP000075880 UP000075920 UP000030765 UP000002320 UP000075885 UP000000311 UP000076408 UP000075883 UP000075903 UP000007062 UP000036403 UP000075840 UP000076407 UP000069272 UP000075902 UP000075882 UP000000673 UP000075886 UP000075884 UP000005205 UP000078492 UP000075900 UP000008237 UP000183832 UP000092461 UP000027135 UP000078541 UP000079169 UP000007755 UP000095301 UP000007266 UP000053097 UP000279307 UP000078540 UP000235965 UP000002358 UP000078542 UP000215335 UP000092462 UP000095300 UP000192223 UP000008792 UP000078200 UP000091820 UP000092444 UP000009046 UP000092445
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K2W2
A0A2A4K2I8
A0A2W1BC49
A0A2H1W5N5
A0A194QE01
A0A0N1IAA5
+ More
A0A3S2M0E9 A0A1S4FW99 Q16M32 A0A182GHY1 A0A023ESG9 A0A1Y1NGL9 A0A1Y1NM71 A0A1Y1NGD6 A0A1Y1NHG5 A0A1Y1NJA7 A0A1Y1NGD0 A0A1Q3FJB8 A0A1Q3FJ43 A0A182JTU0 A0A182IJS5 A0A182VZY7 A0A0C9RUD5 A0A084W717 B0W6D5 A0A182P3V4 E2AUA1 A0A182Y6Q6 A0A182M1T4 A0A182UNQ8 Q5TTX4 A0A0J7KDR9 A0A182HSC6 A0A182XAI6 A0A182FAH2 T1E8U9 A0A182U9H6 A0A182LEB8 W5JAK3 A0A182QJW2 A0A2M3Z129 U5ESB4 A0A2M4BMH5 A0A2M4BM28 A0A2M4BMM0 A0A182NR99 A0A158P2A0 A0A195E4D4 A0A182RQL2 E2C029 A0A1L8DE01 A0A1J1IXH4 A0A2M4AD31 A0A2M4ACT4 A0A1L8DEC7 A0A1L8DDY3 A0A1L8DE06 A0A1B0GKB1 A0A1B0EWL2 A0A067RI22 W8BLL6 A0A195FIT7 A0A2M4ABC1 V5GU37 A0A3Q0J802 F4W6J5 A0A1I8MQ07 A0A1B6CY20 A0A2M4ABD5 D6WZN2 A0A026WF95 A0A1I8NKQ8 A0A195BT61 A0A2J7PTH0 A0A2J7PTH4 K7J059 A0A195D319 A0A232FCH1 A0A1B0DC13 A0A1I8Q5L4 A0A1I8NEP0 A0A1W4WPR9 B4M6U9 A0A146LKT8 A0A0K8SEF7 A0A0A9YHI4 A0A1A9UM78 A0A034W221 A0A0K8U568 A0A336MLN3 A0A1A9WDB0 A0A336M0W0 D3TPB7 E0VCS0 A0A1A9Z8L0 A0A1B6F323 A0A0A1XB07 A0A224XJS0
A0A3S2M0E9 A0A1S4FW99 Q16M32 A0A182GHY1 A0A023ESG9 A0A1Y1NGL9 A0A1Y1NM71 A0A1Y1NGD6 A0A1Y1NHG5 A0A1Y1NJA7 A0A1Y1NGD0 A0A1Q3FJB8 A0A1Q3FJ43 A0A182JTU0 A0A182IJS5 A0A182VZY7 A0A0C9RUD5 A0A084W717 B0W6D5 A0A182P3V4 E2AUA1 A0A182Y6Q6 A0A182M1T4 A0A182UNQ8 Q5TTX4 A0A0J7KDR9 A0A182HSC6 A0A182XAI6 A0A182FAH2 T1E8U9 A0A182U9H6 A0A182LEB8 W5JAK3 A0A182QJW2 A0A2M3Z129 U5ESB4 A0A2M4BMH5 A0A2M4BM28 A0A2M4BMM0 A0A182NR99 A0A158P2A0 A0A195E4D4 A0A182RQL2 E2C029 A0A1L8DE01 A0A1J1IXH4 A0A2M4AD31 A0A2M4ACT4 A0A1L8DEC7 A0A1L8DDY3 A0A1L8DE06 A0A1B0GKB1 A0A1B0EWL2 A0A067RI22 W8BLL6 A0A195FIT7 A0A2M4ABC1 V5GU37 A0A3Q0J802 F4W6J5 A0A1I8MQ07 A0A1B6CY20 A0A2M4ABD5 D6WZN2 A0A026WF95 A0A1I8NKQ8 A0A195BT61 A0A2J7PTH0 A0A2J7PTH4 K7J059 A0A195D319 A0A232FCH1 A0A1B0DC13 A0A1I8Q5L4 A0A1I8NEP0 A0A1W4WPR9 B4M6U9 A0A146LKT8 A0A0K8SEF7 A0A0A9YHI4 A0A1A9UM78 A0A034W221 A0A0K8U568 A0A336MLN3 A0A1A9WDB0 A0A336M0W0 D3TPB7 E0VCS0 A0A1A9Z8L0 A0A1B6F323 A0A0A1XB07 A0A224XJS0
PDB
3ZUX
E-value=9.4286e-08,
Score=131
Ontologies
GO
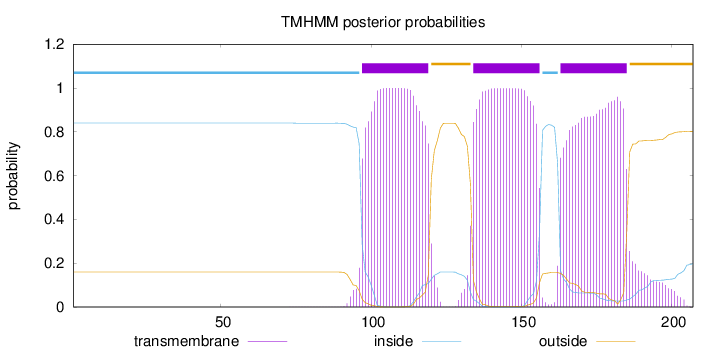
Topology
Length:
207
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.14781
Exp number, first 60 AAs:
8e-05
Total prob of N-in:
0.83985
inside
1 - 96
TMhelix
97 - 119
outside
120 - 133
TMhelix
134 - 156
inside
157 - 162
TMhelix
163 - 185
outside
186 - 207
Population Genetic Test Statistics
Pi
244.814146
Theta
162.963059
Tajima's D
2.150852
CLR
0
CSRT
0.904704764761762
Interpretation
Uncertain
