Pre Gene Modal
BGIBMGA010697
Annotation
putative_transcription_factor_TFIIH-subunit_[Danaus_plexippus]
Full name
Adenylyl cyclase-associated protein
Location in the cell
Extracellular Reliability : 1.367 PlasmaMembrane Reliability : 1.602
Sequence
CDS
ATGGCAGACGACGATGTGCCCTCCGACAGCAGTCTCCTTGTGGTAATAGTGGACACCAATCCCAATCAAAGATACATAATAGAGGACCCCAAAATGCTCACAAACGTCTTGGATGCTGTAGTAGCATTTAGTAACTCCCATTTAATGCAGAAAGCTAGTAATGAACTTGCAGTAATAGGCTGTCATTTTCATAAAAGTGAATACCTGTATCCAAGTCCTGGTAAACCTCTCGATGTAAGACAAATAGATGGACAGTATGAATTATTTACATTAGTTGAGAAGACCATCAAAATGAGATTAGTCAATTTAATCAAAAGTCAACCACAAGAAGAGAAACCCGGAGAATCATTGCTGGCCGGCGCTTTGGCACTGGGACTCTGTTTTATTGCCAGATCTCGACGTAGTGCCATTCCGGGTTTAAGGACTAGCAGCCGTATCCTCGTGGTCACGGGCAGCGCCGACACCGCTGCGCAGTACATCAACTACATGAACGTGTTCTTCACCGCACAGAAGGAGCAAGTCCTGATCGACGTGTGCTCTGTGGACAAACACTTGAGTCTGCTGCAACAGGGTTGCGACATCACGGGCGGCCTGTACCTGAAGATACCGAGCTTGGAGGGCTTGCTGCAGTACTTACTGTGGGTTTTCCTGCCGGAAGCCAATGAGAGAAGTAAACTGGTACTGCCGGGGCGGGGCCGCGTGGACTACCGCGCTGCCTGCTTCTGCCACAGGGAACTGCTCACCATCGGCTACGTCTGCTCGGTTTGCTTGAGTGTTTTCTGCAAATTCAGCCCGATATGTACAACGTGTCACACCGTCTTCAAAATAGGAGGACCACCTATCAAACCCAAGAAGAAGAAAATGAAAGTTTAA
Protein
MADDDVPSDSSLLVVIVDTNPNQRYIIEDPKMLTNVLDAVVAFSNSHLMQKASNELAVIGCHFHKSEYLYPSPGKPLDVRQIDGQYELFTLVEKTIKMRLVNLIKSQPQEEKPGESLLAGALALGLCFIARSRRSAIPGLRTSSRILVVTGSADTAAQYINYMNVFFTAQKEQVLIDVCSVDKHLSLLQQGCDITGGLYLKIPSLEGLLQYLLWVFLPEANERSKLVLPGRGRVDYRAACFCHRELLTIGYVCSVCLSVFCKFSPICTTCHTVFKIGGPPIKPKKKKMKV
Summary
Similarity
Belongs to the CAP family.
Uniprot
A0A2A4K2H9
A0A3S2NTM9
A0A2H1VT08
A0A0N1PHB8
A0A212FDF5
H9JME4
+ More
A0A182JN18 A0A182LZS8 W5JDA3 A0A2M4CG01 A0A182GCT5 Q16LU5 A0A023EMJ2 A0A182GJH6 Q7PNH4 A0A182KEE0 A0A2Y9D3J7 A0A1S4GYI8 A0A2C9GSF2 A0A182UV70 A0A182ULE4 A0A182L038 A0A2M4AVG3 A0A1L8DI76 A0A1S4FWB7 A0A1Q3F218 A0A1B0CYF6 A0A182F2M3 A0A084VBE7 A0A182RCA1 B0VZZ4 A0A1B0FMP1 A0A1A9UTX8 W8BT14 A0A1I8PK45 W8BMP7 A0A182NB42 A0A182VTQ2 U5EXK1 A0A1I8N5G9 A0A034WMR9 A0A1A9WUC5 A0A0L0CG27 A0A0K8V5J7 U4UNS3 N6U406 A0A1J1ISF1 J3JY24 A0A182Y8G5 A0A139WBN9 A0A336LUC4 A0A0K8UR26 A0A182QJ23 A0A1A9XYS8 A0A1L8DIC9 B4MZJ3 A0A0M4E0F1 B4JDF2 Q9VPX4 A0A0M8ZRB5 B4P313 Q8SZ89 A0A1W4VY89 B4Q6N5 B4ID60 B3MN97 A0A3B0K1U0 B3N8C4 Q29LH3 B4KEU6 A0A1Y1L682 A0A310SCP4 A0A026X3B4 A0A1Y1L693 T1GLW4 E2B4L2 A0A087ZUP1 B4GPY6 A0A2A3E839 B4LU35 A0A0K8UDC5 A0A3B0KTT5 A0A158NHC5 F4X076 A0A195F5U4 A0A0K8VKY9 E2AMB7 A0A195D2X6 E9IMW0 A0A151X5R1 A0A151JMR5 A0A232F5S2 A0A1B6LH36 A0A1B6F944 A0A067RSA9 A0A2J7PJR3 A0A182P4P7 K7IU95 A0A3B0JWP6
A0A182JN18 A0A182LZS8 W5JDA3 A0A2M4CG01 A0A182GCT5 Q16LU5 A0A023EMJ2 A0A182GJH6 Q7PNH4 A0A182KEE0 A0A2Y9D3J7 A0A1S4GYI8 A0A2C9GSF2 A0A182UV70 A0A182ULE4 A0A182L038 A0A2M4AVG3 A0A1L8DI76 A0A1S4FWB7 A0A1Q3F218 A0A1B0CYF6 A0A182F2M3 A0A084VBE7 A0A182RCA1 B0VZZ4 A0A1B0FMP1 A0A1A9UTX8 W8BT14 A0A1I8PK45 W8BMP7 A0A182NB42 A0A182VTQ2 U5EXK1 A0A1I8N5G9 A0A034WMR9 A0A1A9WUC5 A0A0L0CG27 A0A0K8V5J7 U4UNS3 N6U406 A0A1J1ISF1 J3JY24 A0A182Y8G5 A0A139WBN9 A0A336LUC4 A0A0K8UR26 A0A182QJ23 A0A1A9XYS8 A0A1L8DIC9 B4MZJ3 A0A0M4E0F1 B4JDF2 Q9VPX4 A0A0M8ZRB5 B4P313 Q8SZ89 A0A1W4VY89 B4Q6N5 B4ID60 B3MN97 A0A3B0K1U0 B3N8C4 Q29LH3 B4KEU6 A0A1Y1L682 A0A310SCP4 A0A026X3B4 A0A1Y1L693 T1GLW4 E2B4L2 A0A087ZUP1 B4GPY6 A0A2A3E839 B4LU35 A0A0K8UDC5 A0A3B0KTT5 A0A158NHC5 F4X076 A0A195F5U4 A0A0K8VKY9 E2AMB7 A0A195D2X6 E9IMW0 A0A151X5R1 A0A151JMR5 A0A232F5S2 A0A1B6LH36 A0A1B6F944 A0A067RSA9 A0A2J7PJR3 A0A182P4P7 K7IU95 A0A3B0JWP6
Pubmed
26354079
22118469
19121390
20920257
23761445
26483478
+ More
17510324 24945155 12364791 20966253 24438588 24495485 25315136 25348373 26108605 23537049 22516182 25244985 18362917 19820115 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085 28004739 24508170 20798317 21347285 21719571 21282665 28648823 24845553 20075255
17510324 24945155 12364791 20966253 24438588 24495485 25315136 25348373 26108605 23537049 22516182 25244985 18362917 19820115 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085 28004739 24508170 20798317 21347285 21719571 21282665 28648823 24845553 20075255
EMBL
NWSH01000220
PCG78259.1
RSAL01000087
RVE48258.1
ODYU01004268
SOQ43960.1
+ More
KQ459941 KPJ19161.1 AGBW02009060 OWR51771.1 BABH01040047 BABH01040048 AXCM01000666 ADMH02001492 ETN62327.1 GGFL01000099 MBW64277.1 JXUM01054805 KQ561837 KXJ77389.1 CH477890 EAT35304.1 GAPW01003051 JAC10547.1 JXUM01012934 KQ560365 KXJ82797.1 AAAB01008964 EAA12181.5 APCN01000743 GGFK01011381 MBW44702.1 GFDF01007999 JAV06085.1 GFDL01013441 JAV21604.1 AJVK01000130 ATLV01006583 KE524368 KFB35291.1 DS231816 EDS37566.1 CCAG010018214 GAMC01006507 JAC00049.1 GAMC01006503 JAC00053.1 GANO01002500 JAB57371.1 GAKP01002106 JAC56846.1 JRES01000438 KNC31172.1 GDHF01018459 JAI33855.1 KB632294 ERL91675.1 APGK01050296 KB741169 ENN73317.1 CVRI01000055 CRL01457.1 BT128148 AEE63109.1 KQ971372 KYB25378.1 UFQS01000115 UFQT01000115 SSW99889.1 SSX20269.1 GDHF01023346 JAI28968.1 AXCN02000644 GFDF01008000 JAV06084.1 CH963920 EDW77778.1 CP012523 ALC38578.1 CH916368 EDW03322.1 AE014134 BT025234 AAF51411.2 ABF17925.1 KQ435885 KOX69540.1 CM000157 EDW87220.1 AY071043 AAL48665.1 CM000361 CM002910 EDX03301.1 KMY87428.1 CH480829 EDW45486.1 CH902620 EDV31054.2 OUUW01000004 SPP79919.1 CH954177 EDV57311.1 CH379060 EAL34072.2 CH933807 EDW12996.1 GEZM01063213 JAV69193.1 KQ761246 OAD57933.1 KK107020 EZA62506.1 GEZM01063214 JAV69192.1 CAQQ02098498 GL445574 EFN89365.1 CH479187 EDW39658.1 KZ288351 PBC27349.1 CH940649 EDW65088.2 GDHF01027675 JAI24639.1 OUUW01000695 SPP90179.1 ADTU01015573 GL888493 EGI60141.1 KQ981768 KYN35843.1 GDHF01012762 JAI39552.1 GL440811 EFN65408.1 KQ976973 KYN06714.1 GL764255 EFZ18109.1 KQ982510 KYQ55639.1 KQ978899 KYN27619.1 NNAY01000940 OXU25813.1 GEBQ01017047 JAT22930.1 GECZ01023020 JAS46749.1 KK852498 KDR22694.1 NEVH01024946 PNF16573.1 AAZX01002622 SPP79920.1
KQ459941 KPJ19161.1 AGBW02009060 OWR51771.1 BABH01040047 BABH01040048 AXCM01000666 ADMH02001492 ETN62327.1 GGFL01000099 MBW64277.1 JXUM01054805 KQ561837 KXJ77389.1 CH477890 EAT35304.1 GAPW01003051 JAC10547.1 JXUM01012934 KQ560365 KXJ82797.1 AAAB01008964 EAA12181.5 APCN01000743 GGFK01011381 MBW44702.1 GFDF01007999 JAV06085.1 GFDL01013441 JAV21604.1 AJVK01000130 ATLV01006583 KE524368 KFB35291.1 DS231816 EDS37566.1 CCAG010018214 GAMC01006507 JAC00049.1 GAMC01006503 JAC00053.1 GANO01002500 JAB57371.1 GAKP01002106 JAC56846.1 JRES01000438 KNC31172.1 GDHF01018459 JAI33855.1 KB632294 ERL91675.1 APGK01050296 KB741169 ENN73317.1 CVRI01000055 CRL01457.1 BT128148 AEE63109.1 KQ971372 KYB25378.1 UFQS01000115 UFQT01000115 SSW99889.1 SSX20269.1 GDHF01023346 JAI28968.1 AXCN02000644 GFDF01008000 JAV06084.1 CH963920 EDW77778.1 CP012523 ALC38578.1 CH916368 EDW03322.1 AE014134 BT025234 AAF51411.2 ABF17925.1 KQ435885 KOX69540.1 CM000157 EDW87220.1 AY071043 AAL48665.1 CM000361 CM002910 EDX03301.1 KMY87428.1 CH480829 EDW45486.1 CH902620 EDV31054.2 OUUW01000004 SPP79919.1 CH954177 EDV57311.1 CH379060 EAL34072.2 CH933807 EDW12996.1 GEZM01063213 JAV69193.1 KQ761246 OAD57933.1 KK107020 EZA62506.1 GEZM01063214 JAV69192.1 CAQQ02098498 GL445574 EFN89365.1 CH479187 EDW39658.1 KZ288351 PBC27349.1 CH940649 EDW65088.2 GDHF01027675 JAI24639.1 OUUW01000695 SPP90179.1 ADTU01015573 GL888493 EGI60141.1 KQ981768 KYN35843.1 GDHF01012762 JAI39552.1 GL440811 EFN65408.1 KQ976973 KYN06714.1 GL764255 EFZ18109.1 KQ982510 KYQ55639.1 KQ978899 KYN27619.1 NNAY01000940 OXU25813.1 GEBQ01017047 JAT22930.1 GECZ01023020 JAS46749.1 KK852498 KDR22694.1 NEVH01024946 PNF16573.1 AAZX01002622 SPP79920.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000007151
UP000005204
UP000075880
+ More
UP000075883 UP000000673 UP000069940 UP000249989 UP000008820 UP000007062 UP000075881 UP000076407 UP000075840 UP000075903 UP000075902 UP000075882 UP000092462 UP000069272 UP000030765 UP000075900 UP000002320 UP000092444 UP000078200 UP000095300 UP000075884 UP000075920 UP000095301 UP000091820 UP000037069 UP000030742 UP000019118 UP000183832 UP000076408 UP000007266 UP000075886 UP000092443 UP000007798 UP000092553 UP000001070 UP000000803 UP000053105 UP000002282 UP000192221 UP000000304 UP000001292 UP000007801 UP000268350 UP000008711 UP000001819 UP000009192 UP000053097 UP000015102 UP000008237 UP000005203 UP000008744 UP000242457 UP000008792 UP000005205 UP000007755 UP000078541 UP000000311 UP000078542 UP000075809 UP000078492 UP000215335 UP000027135 UP000235965 UP000075885 UP000002358
UP000075883 UP000000673 UP000069940 UP000249989 UP000008820 UP000007062 UP000075881 UP000076407 UP000075840 UP000075903 UP000075902 UP000075882 UP000092462 UP000069272 UP000030765 UP000075900 UP000002320 UP000092444 UP000078200 UP000095300 UP000075884 UP000075920 UP000095301 UP000091820 UP000037069 UP000030742 UP000019118 UP000183832 UP000076408 UP000007266 UP000075886 UP000092443 UP000007798 UP000092553 UP000001070 UP000000803 UP000053105 UP000002282 UP000192221 UP000000304 UP000001292 UP000007801 UP000268350 UP000008711 UP000001819 UP000009192 UP000053097 UP000015102 UP000008237 UP000005203 UP000008744 UP000242457 UP000008792 UP000005205 UP000007755 UP000078541 UP000000311 UP000078542 UP000075809 UP000078492 UP000215335 UP000027135 UP000235965 UP000075885 UP000002358
Interpro
IPR004600
TFIIH_Tfb4/GTF2H3
+ More
IPR036465 vWFA_dom_sf
IPR006020 PTB/PI_dom
IPR011993 PH-like_dom_sf
IPR006599 CARP_motif
IPR036222 CAP_N_sf
IPR016098 CAP/MinC_C
IPR013992 Adenylate_cyclase-assoc_CAP_N
IPR036223 CAP_C_sf
IPR001837 Adenylate_cyclase-assoc_CAP
IPR028417 CAP_CS_C
IPR013912 Adenylate_cyclase-assoc_CAP_C
IPR017901 C-CAP_CF_C-like
IPR018106 CAP_CS_N
IPR036465 vWFA_dom_sf
IPR006020 PTB/PI_dom
IPR011993 PH-like_dom_sf
IPR006599 CARP_motif
IPR036222 CAP_N_sf
IPR016098 CAP/MinC_C
IPR013992 Adenylate_cyclase-assoc_CAP_N
IPR036223 CAP_C_sf
IPR001837 Adenylate_cyclase-assoc_CAP
IPR028417 CAP_CS_C
IPR013912 Adenylate_cyclase-assoc_CAP_C
IPR017901 C-CAP_CF_C-like
IPR018106 CAP_CS_N
Gene 3D
ProteinModelPortal
A0A2A4K2H9
A0A3S2NTM9
A0A2H1VT08
A0A0N1PHB8
A0A212FDF5
H9JME4
+ More
A0A182JN18 A0A182LZS8 W5JDA3 A0A2M4CG01 A0A182GCT5 Q16LU5 A0A023EMJ2 A0A182GJH6 Q7PNH4 A0A182KEE0 A0A2Y9D3J7 A0A1S4GYI8 A0A2C9GSF2 A0A182UV70 A0A182ULE4 A0A182L038 A0A2M4AVG3 A0A1L8DI76 A0A1S4FWB7 A0A1Q3F218 A0A1B0CYF6 A0A182F2M3 A0A084VBE7 A0A182RCA1 B0VZZ4 A0A1B0FMP1 A0A1A9UTX8 W8BT14 A0A1I8PK45 W8BMP7 A0A182NB42 A0A182VTQ2 U5EXK1 A0A1I8N5G9 A0A034WMR9 A0A1A9WUC5 A0A0L0CG27 A0A0K8V5J7 U4UNS3 N6U406 A0A1J1ISF1 J3JY24 A0A182Y8G5 A0A139WBN9 A0A336LUC4 A0A0K8UR26 A0A182QJ23 A0A1A9XYS8 A0A1L8DIC9 B4MZJ3 A0A0M4E0F1 B4JDF2 Q9VPX4 A0A0M8ZRB5 B4P313 Q8SZ89 A0A1W4VY89 B4Q6N5 B4ID60 B3MN97 A0A3B0K1U0 B3N8C4 Q29LH3 B4KEU6 A0A1Y1L682 A0A310SCP4 A0A026X3B4 A0A1Y1L693 T1GLW4 E2B4L2 A0A087ZUP1 B4GPY6 A0A2A3E839 B4LU35 A0A0K8UDC5 A0A3B0KTT5 A0A158NHC5 F4X076 A0A195F5U4 A0A0K8VKY9 E2AMB7 A0A195D2X6 E9IMW0 A0A151X5R1 A0A151JMR5 A0A232F5S2 A0A1B6LH36 A0A1B6F944 A0A067RSA9 A0A2J7PJR3 A0A182P4P7 K7IU95 A0A3B0JWP6
A0A182JN18 A0A182LZS8 W5JDA3 A0A2M4CG01 A0A182GCT5 Q16LU5 A0A023EMJ2 A0A182GJH6 Q7PNH4 A0A182KEE0 A0A2Y9D3J7 A0A1S4GYI8 A0A2C9GSF2 A0A182UV70 A0A182ULE4 A0A182L038 A0A2M4AVG3 A0A1L8DI76 A0A1S4FWB7 A0A1Q3F218 A0A1B0CYF6 A0A182F2M3 A0A084VBE7 A0A182RCA1 B0VZZ4 A0A1B0FMP1 A0A1A9UTX8 W8BT14 A0A1I8PK45 W8BMP7 A0A182NB42 A0A182VTQ2 U5EXK1 A0A1I8N5G9 A0A034WMR9 A0A1A9WUC5 A0A0L0CG27 A0A0K8V5J7 U4UNS3 N6U406 A0A1J1ISF1 J3JY24 A0A182Y8G5 A0A139WBN9 A0A336LUC4 A0A0K8UR26 A0A182QJ23 A0A1A9XYS8 A0A1L8DIC9 B4MZJ3 A0A0M4E0F1 B4JDF2 Q9VPX4 A0A0M8ZRB5 B4P313 Q8SZ89 A0A1W4VY89 B4Q6N5 B4ID60 B3MN97 A0A3B0K1U0 B3N8C4 Q29LH3 B4KEU6 A0A1Y1L682 A0A310SCP4 A0A026X3B4 A0A1Y1L693 T1GLW4 E2B4L2 A0A087ZUP1 B4GPY6 A0A2A3E839 B4LU35 A0A0K8UDC5 A0A3B0KTT5 A0A158NHC5 F4X076 A0A195F5U4 A0A0K8VKY9 E2AMB7 A0A195D2X6 E9IMW0 A0A151X5R1 A0A151JMR5 A0A232F5S2 A0A1B6LH36 A0A1B6F944 A0A067RSA9 A0A2J7PJR3 A0A182P4P7 K7IU95 A0A3B0JWP6
PDB
6O9M
E-value=1.81409e-65,
Score=631
Ontologies
PATHWAY
GO
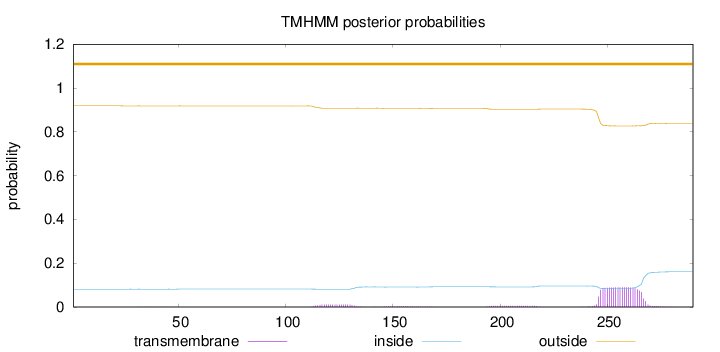
Topology
Length:
290
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.29408
Exp number, first 60 AAs:
0.00746
Total prob of N-in:
0.08140
outside
1 - 290
Population Genetic Test Statistics
Pi
238.11605
Theta
177.343363
Tajima's D
1.188428
CLR
0.384356
CSRT
0.708214589270536
Interpretation
Uncertain
