Gene
KWMTBOMO15645
Pre Gene Modal
BGIBMGA010696
Annotation
PREDICTED:_uncharacterized_protein_LOC105202492_[Solenopsis_invicta]
Transcription factor
Location in the cell
Nuclear Reliability : 3.971
Sequence
CDS
ATGCCTAATAAATACAAGAGAAAGGCAGTGGCTGTGAGAGGAAATTGGTCGGAAGAATCACTAAAAGCAGCTATTAATGCTGTGAAAAATGATGGATTATCCGTACGCGCGGCTAGTCTCCAGTATCAGATACCTAGAAAGGCATTAGAACGAAAGCTAAAGAAGAATGACGACAAGAAAGGCCCTATGGGACCACCTACTTTATTCGGAGAACAAAATGAGAAAAAATTGGTTGCTCATATTAAAACAATATTTAACAAAGACAATCAGAAAGCAGGATATGACTGGTTGCAGTTGTTCTTGAGCAGAAATACTGATATTGCACTTAGGAAGTCAGAAGGAGTTTCTTTGGCTAGAGTCAATGGCATGAATAGATCAGAAGTTAATGCATATTTTAAATTGCTTGAAAGCGTCCTTATCCATGAGAATGAGATGCTACCTCCTAACTGCGTATTTAACATGGATGAATCTGGCCTACAGTTAAATAGTCGTCCGGGGCACGTGCTTGCTCAAAAAGGTTCTAAAGCTGTTTCCACAGTGACATCAACTGAAAAAGGGGAGACAATAACCATTATAGCCTGCTGCAATGCAGAAGGCACGTTCTTGCCACCTGCCTGTATCATGAAAGGAAAAAATAAAAAGCCTGAGTTTGAAGACGGAATGCCTCCAGGATCAAAACTATTCATGTCACAAAAATCAGCATATATAATATCAGAAATATTTTTAGAATGGTTAAAAACACACTTTGTTCCAAGGAAACCAGCCGGGAAAGTTGTCCTTCTGTTAGACGGCCACTCAACTCATTCCAATTCTGTGGAAATGTTGGAATATGCCATTCAGAATGACATTATTTTGGTATCAATGCCTAGTCACACGTCTCATTATCTCCAGCCCTTAGATCGTTCTGTTTTTAAACCACTAAAACATCACTTTTATGAACAGTCTAGACTTTGGTTAATCCACAACCCAGGACGTCGAATATCGAGACTTTCTTTCGGAGTTTTGCTTAGCAAGGCTTGGGGAAAGGCAGCATCGTCGGAAAATGCAATTGCTGGTTTTAAAGCGACTGGAGTGTATCTTCTTAACCCAGAAGTGGTGCCAGATTATGCGTTTTTTCAAGAATCTACAGGTGACTCGACGAATACTTCTGTTACACAAGTGCTTTCACATTGTAAATCTGCTGTGGACAATGAGATACCAGGTTGTTCTTATGAACCAATAATAGATGAAACAAAATATAGCTCTGAAACCGTGTCTACAAAATCCTCCGTTAAAACTGCCACGATGTCTAATATGTCTCACCCGAGTACTCCGAAAGAAGCTTCGTTCAGTAAGAATCCAGACTACACCCCATCTCGAATTTTACAGGATATATCACCTATTCCACAAAAACTGATTCAGGTGCGTAAACGAGCAAAGCAAGTTGGAACACTATTGACTTCGGAAGAACACATCAAAACTAGAAAAAATGCTGCAGAAAAAAAGGAAAACGTAAAAAAATGTGTCAAAACAAAGCAGGAAAAAAAAACTGTTCTATGTATAAAAAGAAAAGCCATTCGAAGACTATCTGAGAATAGTGACGAAATTCCATTAAAAATATTGTCAGTTAAGAATGAGAAAAGAAAGCTAGATGAAAAGGATAACTGTCGTGGGTGTGGTGAAAATTACTACGAAACACTTCTCATTGAAGACTGGCTTCAATGTACAGTCTGTCAGTATTGGGTTCACGAACACTGCACTGAATTTGAAACGACGTGCTCCAGATGTGGCGAAGAGGAGAAAACAAGACTTAAAAATAGAAAATGA
Protein
MPNKYKRKAVAVRGNWSEESLKAAINAVKNDGLSVRAASLQYQIPRKALERKLKKNDDKKGPMGPPTLFGEQNEKKLVAHIKTIFNKDNQKAGYDWLQLFLSRNTDIALRKSEGVSLARVNGMNRSEVNAYFKLLESVLIHENEMLPPNCVFNMDESGLQLNSRPGHVLAQKGSKAVSTVTSTEKGETITIIACCNAEGTFLPPACIMKGKNKKPEFEDGMPPGSKLFMSQKSAYIISEIFLEWLKTHFVPRKPAGKVVLLLDGHSTHSNSVEMLEYAIQNDIILVSMPSHTSHYLQPLDRSVFKPLKHHFYEQSRLWLIHNPGRRISRLSFGVLLSKAWGKAASSENAIAGFKATGVYLLNPEVVPDYAFFQESTGDSTNTSVTQVLSHCKSAVDNEIPGCSYEPIIDETKYSSETVSTKSSVKTATMSNMSHPSTPKEASFSKNPDYTPSRILQDISPIPQKLIQVRKRAKQVGTLLTSEEHIKTRKNAAEKKENVKKCVKTKQEKKTVLCIKRKAIRRLSENSDEIPLKILSVKNEKRKLDEKDNCRGCGENYYETLLIEDWLQCTVCQYWVHEHCTEFETTCSRCGEEEKTRLKNRK
Summary
Uniprot
H9JME3
A0A2H1X459
A0A3L8DSU8
A0A1Y1JVK5
A0A212EU96
J9KB99
+ More
A0A146KT86 A0A2H8TEI8 A0A2H1VWF9 A0A1Y1LFM2 J9LG75 J9KKA5 A0A1Y1MUL7 A0A3L8DD48 T1EQM1 A0A1S3D1W0 A0A1Y1NGN4 A0A1Y1NJC0 A0A1B6ISM7 A0A1Y1NBD5 A0A2A4J258 A0A2A4J1M3 A0A2H1VTA0 J9JKJ5 H9JME9 A0A2A4J6W1 A0A194RH46 A0A2A4K4E0 A0A1W4XA29 A0A2W1BH88 A0A2H9T3C2
A0A146KT86 A0A2H8TEI8 A0A2H1VWF9 A0A1Y1LFM2 J9LG75 J9KKA5 A0A1Y1MUL7 A0A3L8DD48 T1EQM1 A0A1S3D1W0 A0A1Y1NGN4 A0A1Y1NJC0 A0A1B6ISM7 A0A1Y1NBD5 A0A2A4J258 A0A2A4J1M3 A0A2H1VTA0 J9JKJ5 H9JME9 A0A2A4J6W1 A0A194RH46 A0A2A4K4E0 A0A1W4XA29 A0A2W1BH88 A0A2H9T3C2
EMBL
BABH01040047
ODYU01013314
SOQ60022.1
QOIP01000004
RLU23500.1
GEZM01099403
+ More
GEZM01099402 JAV53344.1 AGBW02012433 OWR45073.1 ABLF02022753 GDHC01018928 JAP99700.1 GFXV01000711 MBW12516.1 ODYU01004859 SOQ45169.1 GEZM01058773 JAV71681.1 ABLF02009840 ABLF02016204 ABLF02006839 GEZM01026324 JAV87047.1 QOIP01000010 RLU18063.1 AMQM01000642 KB096324 ESO06516.1 GEZM01002997 JAV97001.1 GEZM01002998 JAV97000.1 GECU01017774 JAS89932.1 GEZM01007306 JAV95254.1 NWSH01003844 PCG65768.1 PCG65769.1 ODYU01004314 SOQ44057.1 ABLF02004627 ABLF02004628 ABLF02034930 BABH01016912 NWSH01002933 PCG67263.1 PCG67264.1 KQ460205 KPJ16902.1 NWSH01000193 PCG78522.1 KZ150051 PZC74412.1 NSIT01000429 PJE77687.1
GEZM01099402 JAV53344.1 AGBW02012433 OWR45073.1 ABLF02022753 GDHC01018928 JAP99700.1 GFXV01000711 MBW12516.1 ODYU01004859 SOQ45169.1 GEZM01058773 JAV71681.1 ABLF02009840 ABLF02016204 ABLF02006839 GEZM01026324 JAV87047.1 QOIP01000010 RLU18063.1 AMQM01000642 KB096324 ESO06516.1 GEZM01002997 JAV97001.1 GEZM01002998 JAV97000.1 GECU01017774 JAS89932.1 GEZM01007306 JAV95254.1 NWSH01003844 PCG65768.1 PCG65769.1 ODYU01004314 SOQ44057.1 ABLF02004627 ABLF02004628 ABLF02034930 BABH01016912 NWSH01002933 PCG67263.1 PCG67264.1 KQ460205 KPJ16902.1 NWSH01000193 PCG78522.1 KZ150051 PZC74412.1 NSIT01000429 PJE77687.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9JME3
A0A2H1X459
A0A3L8DSU8
A0A1Y1JVK5
A0A212EU96
J9KB99
+ More
A0A146KT86 A0A2H8TEI8 A0A2H1VWF9 A0A1Y1LFM2 J9LG75 J9KKA5 A0A1Y1MUL7 A0A3L8DD48 T1EQM1 A0A1S3D1W0 A0A1Y1NGN4 A0A1Y1NJC0 A0A1B6ISM7 A0A1Y1NBD5 A0A2A4J258 A0A2A4J1M3 A0A2H1VTA0 J9JKJ5 H9JME9 A0A2A4J6W1 A0A194RH46 A0A2A4K4E0 A0A1W4XA29 A0A2W1BH88 A0A2H9T3C2
A0A146KT86 A0A2H8TEI8 A0A2H1VWF9 A0A1Y1LFM2 J9LG75 J9KKA5 A0A1Y1MUL7 A0A3L8DD48 T1EQM1 A0A1S3D1W0 A0A1Y1NGN4 A0A1Y1NJC0 A0A1B6ISM7 A0A1Y1NBD5 A0A2A4J258 A0A2A4J1M3 A0A2H1VTA0 J9JKJ5 H9JME9 A0A2A4J6W1 A0A194RH46 A0A2A4K4E0 A0A1W4XA29 A0A2W1BH88 A0A2H9T3C2
Ontologies
GO
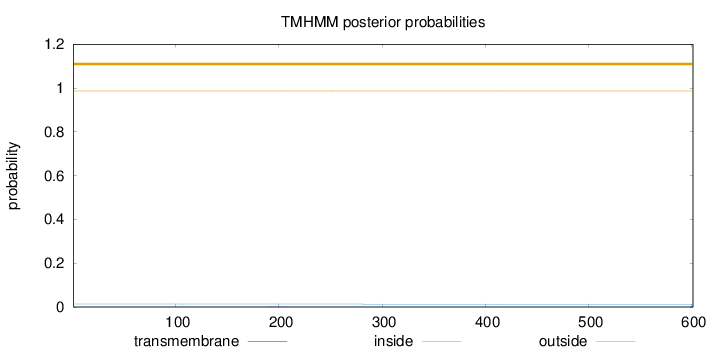
Topology
Subcellular location
Nucleus
Length:
601
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00541999999999999
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.01337
outside
1 - 601
Population Genetic Test Statistics
Pi
581.848816
Theta
228.310752
Tajima's D
4.902524
CLR
0
CSRT
0.999950002499875
Interpretation
Uncertain
