Gene
KWMTBOMO15638
Annotation
PREDICTED:_uncharacterized_protein_LOC105842659_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.089
Sequence
CDS
ATGCGCAAGACCCTAGAAAACTGGGATGGAGGCATAACCATAGGTGGTGTCAAAGTGACGAACCTGCGCTACGCAGATGACACCACTCTGCTTGCCACAACAGAAGCAGAGATGACAGAGCTTCTGAACAGGATGGAGCAGATAGGGTTGGAGATGGGACTCGCCATCAACAGATCCAAAACTAAGATCATGGTCATTGATCGCACAAAAAAGTTGGAGCTTAGTGGAACACTAAACCTTGAGCTTGTTGACAACTTTATTTACCTAGGCTCCAACATTAACAATACTGGATCCAGCGAACTTGAAGTACGACGGCGTATTGGAATGGCAAAGGGAGCCATGACCCAGCTCGGTAAAATCTGGAAGGACCATAACATTACCCAGAAAACAAAGACAAAACTGGTGCTCACTCTGGTCTTTTCCATTTTTATATACGGAACGGAGACCTGGACCCTTAAAGCAGCTGATCGCAAACGCGTGGATGCTTTTGAGATGTGGTGCTAG
Protein
MRKTLENWDGGITIGGVKVTNLRYADDTTLLATTEAEMTELLNRMEQIGLEMGLAINRSKTKIMVIDRTKKLELSGTLNLELVDNFIYLGSNINNTGSSELEVRRRIGMAKGAMTQLGKIWKDHNITQKTKTKLVLTLVFSIFIYGTETWTLKAADRKRVDAFEMWC
Summary
Uniprot
D7F168
D7F163
A0A2W1BGD9
D7F172
D7F161
D7F162
+ More
A0A2H1VU13 D7F167 D7F175 A0A3S2L0R7 A0A2W1BNH4 A0A1J5WD64 A0A2W1B3T2 A0A2G8JE23 D5LB39 A0A2G8JG39 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 H2YWZ5 A0A3Q1M7G0 A0A3Q1LIK6 A0A3Q1LVS6 F6R5D0 A0A3Q1M5E7 A0A3Q1MDY8 A0A3Q1MTQ8 A0A3S3P648 A0A3Q1MZJ0 A0A3Q1LXF5 A0A3Q1LQN1 A0A3Q1N3Y9 A0A3Q1LNR9 W4Z0F5 A0A3Q1MFW2 A0A3Q1MP18 A0A3Q1MMS2 A0A3Q1MJK8 A0A3Q1MGV6 A0A3Q1LK56 A0A3Q1MDZ3 A0A3Q1MFU8 A0A3Q1MIT2 A0A2H6KKM9 A0A3Q1ML12 A0A3Q1NJH7 A0A3Q1MGA3 A0A3Q1MWB3 A0A146L0Z1 A0A3Q1LT63 A0A061BKT1 A0A3Q1N9K5 A0A3Q1LWH0 A0A3Q1MCX3 A0A3Q1ML90 A0A0A9XXL9 A0A3Q1LSI8 A0A3Q1MQ02 A0A3Q1N2X2 A0A3Q1LKN4 A0A3Q1MSR0 A0A3S3NR17 A0A2G8KEY9 A0A3S3RLW1 A0A3Q1LQF3 A0A3Q1MVG9 A0A3Q1NGZ5 A0A3Q1LN91 A0A3Q1LX64 A0A3Q1LGI2 W5MXL2 A0A3Q1MBA9 A0A3Q1LXQ5 A0A3Q1M427 W5PYP3 A0A3Q1LGI4 A0A3Q1M4E7 W5NGM3 W4YPU3 A0A3Q1MJR3 A0A3Q1MBV2 A0A3Q1LKL4 A0A0J7NYI3 A0A3Q1MVY4 A0A3Q1M6I4 A0A3Q1MLZ9 A0A3Q1M5A3 A0A3Q1MN71 A0A3Q1M3R4 A0A3Q1M1T7 A0A2G8KGL8 A0A3Q1MNQ2 A0A3Q1LR18 A0A3Q1MCA0 D5LB38 A0A3Q1MLI2 A0A3Q1NL67 A0A3Q1LIJ6
A0A2H1VU13 D7F167 D7F175 A0A3S2L0R7 A0A2W1BNH4 A0A1J5WD64 A0A2W1B3T2 A0A2G8JE23 D5LB39 A0A2G8JG39 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 H2YWZ5 A0A3Q1M7G0 A0A3Q1LIK6 A0A3Q1LVS6 F6R5D0 A0A3Q1M5E7 A0A3Q1MDY8 A0A3Q1MTQ8 A0A3S3P648 A0A3Q1MZJ0 A0A3Q1LXF5 A0A3Q1LQN1 A0A3Q1N3Y9 A0A3Q1LNR9 W4Z0F5 A0A3Q1MFW2 A0A3Q1MP18 A0A3Q1MMS2 A0A3Q1MJK8 A0A3Q1MGV6 A0A3Q1LK56 A0A3Q1MDZ3 A0A3Q1MFU8 A0A3Q1MIT2 A0A2H6KKM9 A0A3Q1ML12 A0A3Q1NJH7 A0A3Q1MGA3 A0A3Q1MWB3 A0A146L0Z1 A0A3Q1LT63 A0A061BKT1 A0A3Q1N9K5 A0A3Q1LWH0 A0A3Q1MCX3 A0A3Q1ML90 A0A0A9XXL9 A0A3Q1LSI8 A0A3Q1MQ02 A0A3Q1N2X2 A0A3Q1LKN4 A0A3Q1MSR0 A0A3S3NR17 A0A2G8KEY9 A0A3S3RLW1 A0A3Q1LQF3 A0A3Q1MVG9 A0A3Q1NGZ5 A0A3Q1LN91 A0A3Q1LX64 A0A3Q1LGI2 W5MXL2 A0A3Q1MBA9 A0A3Q1LXQ5 A0A3Q1M427 W5PYP3 A0A3Q1LGI4 A0A3Q1M4E7 W5NGM3 W4YPU3 A0A3Q1MJR3 A0A3Q1MBV2 A0A3Q1LKL4 A0A0J7NYI3 A0A3Q1MVY4 A0A3Q1M6I4 A0A3Q1MLZ9 A0A3Q1M5A3 A0A3Q1MN71 A0A3Q1M3R4 A0A3Q1M1T7 A0A2G8KGL8 A0A3Q1MNQ2 A0A3Q1LR18 A0A3Q1MCA0 D5LB38 A0A3Q1MLI2 A0A3Q1NL67 A0A3Q1LIJ6
EMBL
FJ265553
ADI61821.1
FJ265548
ADI61816.1
KZ150203
PZC72267.1
+ More
FJ265557 ADI61825.1 FJ265546 ADI61814.1 FJ265547 ADI61815.1 ODYU01004401 SOQ44236.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 RSAL01000289 RVE42931.1 KZ150025 PZC74827.1 MKPU01000878 OIR55924.1 KZ150363 PZC71182.1 MRZV01002322 PIK33998.1 GU815090 ADF18553.1 MRZV01002096 PIK34710.1 MRZV01000196 PIK55924.1 MRZV01001207 PIK39730.1 AHAT01022938 NCKU01004395 RWS06003.1 AAGJ04089624 BDSA01000081 GBE63537.1 GDHC01017214 JAQ01415.1 LK054988 CDR71535.1 GBHO01020011 GDHC01013600 JAG23593.1 JAQ05029.1 NCKU01012182 RWS00163.1 MRZV01000636 PIK46566.1 NCKU01008836 RWS01655.1 AHAT01017596 AMGL01118292 AMGL01118293 AHAT01008485 AAGJ04055194 LBMM01000801 KMQ97460.1 MRZV01000599 PIK47151.1 GU815089 ADF18552.1
FJ265557 ADI61825.1 FJ265546 ADI61814.1 FJ265547 ADI61815.1 ODYU01004401 SOQ44236.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 RSAL01000289 RVE42931.1 KZ150025 PZC74827.1 MKPU01000878 OIR55924.1 KZ150363 PZC71182.1 MRZV01002322 PIK33998.1 GU815090 ADF18553.1 MRZV01002096 PIK34710.1 MRZV01000196 PIK55924.1 MRZV01001207 PIK39730.1 AHAT01022938 NCKU01004395 RWS06003.1 AAGJ04089624 BDSA01000081 GBE63537.1 GDHC01017214 JAQ01415.1 LK054988 CDR71535.1 GBHO01020011 GDHC01013600 JAG23593.1 JAQ05029.1 NCKU01012182 RWS00163.1 MRZV01000636 PIK46566.1 NCKU01008836 RWS01655.1 AHAT01017596 AMGL01118292 AMGL01118293 AHAT01008485 AAGJ04055194 LBMM01000801 KMQ97460.1 MRZV01000599 PIK47151.1 GU815089 ADF18552.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D7F168
D7F163
A0A2W1BGD9
D7F172
D7F161
D7F162
+ More
A0A2H1VU13 D7F167 D7F175 A0A3S2L0R7 A0A2W1BNH4 A0A1J5WD64 A0A2W1B3T2 A0A2G8JE23 D5LB39 A0A2G8JG39 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 H2YWZ5 A0A3Q1M7G0 A0A3Q1LIK6 A0A3Q1LVS6 F6R5D0 A0A3Q1M5E7 A0A3Q1MDY8 A0A3Q1MTQ8 A0A3S3P648 A0A3Q1MZJ0 A0A3Q1LXF5 A0A3Q1LQN1 A0A3Q1N3Y9 A0A3Q1LNR9 W4Z0F5 A0A3Q1MFW2 A0A3Q1MP18 A0A3Q1MMS2 A0A3Q1MJK8 A0A3Q1MGV6 A0A3Q1LK56 A0A3Q1MDZ3 A0A3Q1MFU8 A0A3Q1MIT2 A0A2H6KKM9 A0A3Q1ML12 A0A3Q1NJH7 A0A3Q1MGA3 A0A3Q1MWB3 A0A146L0Z1 A0A3Q1LT63 A0A061BKT1 A0A3Q1N9K5 A0A3Q1LWH0 A0A3Q1MCX3 A0A3Q1ML90 A0A0A9XXL9 A0A3Q1LSI8 A0A3Q1MQ02 A0A3Q1N2X2 A0A3Q1LKN4 A0A3Q1MSR0 A0A3S3NR17 A0A2G8KEY9 A0A3S3RLW1 A0A3Q1LQF3 A0A3Q1MVG9 A0A3Q1NGZ5 A0A3Q1LN91 A0A3Q1LX64 A0A3Q1LGI2 W5MXL2 A0A3Q1MBA9 A0A3Q1LXQ5 A0A3Q1M427 W5PYP3 A0A3Q1LGI4 A0A3Q1M4E7 W5NGM3 W4YPU3 A0A3Q1MJR3 A0A3Q1MBV2 A0A3Q1LKL4 A0A0J7NYI3 A0A3Q1MVY4 A0A3Q1M6I4 A0A3Q1MLZ9 A0A3Q1M5A3 A0A3Q1MN71 A0A3Q1M3R4 A0A3Q1M1T7 A0A2G8KGL8 A0A3Q1MNQ2 A0A3Q1LR18 A0A3Q1MCA0 D5LB38 A0A3Q1MLI2 A0A3Q1NL67 A0A3Q1LIJ6
A0A2H1VU13 D7F167 D7F175 A0A3S2L0R7 A0A2W1BNH4 A0A1J5WD64 A0A2W1B3T2 A0A2G8JE23 D5LB39 A0A2G8JG39 A0A2G8L6P4 A0A2G8JVI4 W5NMM5 H2YWZ5 A0A3Q1M7G0 A0A3Q1LIK6 A0A3Q1LVS6 F6R5D0 A0A3Q1M5E7 A0A3Q1MDY8 A0A3Q1MTQ8 A0A3S3P648 A0A3Q1MZJ0 A0A3Q1LXF5 A0A3Q1LQN1 A0A3Q1N3Y9 A0A3Q1LNR9 W4Z0F5 A0A3Q1MFW2 A0A3Q1MP18 A0A3Q1MMS2 A0A3Q1MJK8 A0A3Q1MGV6 A0A3Q1LK56 A0A3Q1MDZ3 A0A3Q1MFU8 A0A3Q1MIT2 A0A2H6KKM9 A0A3Q1ML12 A0A3Q1NJH7 A0A3Q1MGA3 A0A3Q1MWB3 A0A146L0Z1 A0A3Q1LT63 A0A061BKT1 A0A3Q1N9K5 A0A3Q1LWH0 A0A3Q1MCX3 A0A3Q1ML90 A0A0A9XXL9 A0A3Q1LSI8 A0A3Q1MQ02 A0A3Q1N2X2 A0A3Q1LKN4 A0A3Q1MSR0 A0A3S3NR17 A0A2G8KEY9 A0A3S3RLW1 A0A3Q1LQF3 A0A3Q1MVG9 A0A3Q1NGZ5 A0A3Q1LN91 A0A3Q1LX64 A0A3Q1LGI2 W5MXL2 A0A3Q1MBA9 A0A3Q1LXQ5 A0A3Q1M427 W5PYP3 A0A3Q1LGI4 A0A3Q1M4E7 W5NGM3 W4YPU3 A0A3Q1MJR3 A0A3Q1MBV2 A0A3Q1LKL4 A0A0J7NYI3 A0A3Q1MVY4 A0A3Q1M6I4 A0A3Q1MLZ9 A0A3Q1M5A3 A0A3Q1MN71 A0A3Q1M3R4 A0A3Q1M1T7 A0A2G8KGL8 A0A3Q1MNQ2 A0A3Q1LR18 A0A3Q1MCA0 D5LB38 A0A3Q1MLI2 A0A3Q1NL67 A0A3Q1LIJ6
Ontologies
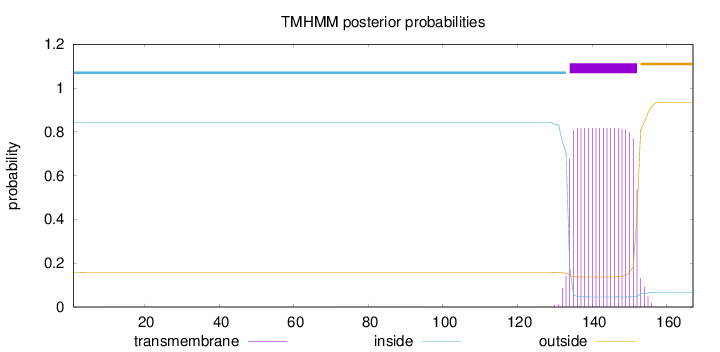
Topology
Length:
167
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.56111
Exp number, first 60 AAs:
0.00308
Total prob of N-in:
0.84277
inside
1 - 133
TMhelix
134 - 152
outside
153 - 167
Population Genetic Test Statistics
Pi
270.858541
Theta
139.69194
Tajima's D
2.950554
CLR
0
CSRT
0.977001149942503
Interpretation
Uncertain
