Gene
KWMTBOMO15635
Pre Gene Modal
BGIBMGA010702
Annotation
PREDICTED:_uncharacterized_protein_LOC105842267_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.282
Sequence
CDS
ATGCCTCGAAAATATATAAGGAAATGTCCCGATCGGAAAGTCTGTACTCAAGAGCAAATTGATAAAGCCAAGCAGTTGATTGGAGAAGGAAAATCGAAACGAGCAGCAGCAGAAATTGTTGGGATCAATGAGAGTACTTTGAGAAAACGTTTAAAACTCAACTCCACTGCAACATCAATGGGGAGATATCAACAAACATTTACACGTGAACAAGAGGAAGAAATTTACAATCATTGCAAATCTAGTGACGAGAGGTTCTACGGTCTTACTTTGAATACACTGAGAAAATTAGTTTACGAATTTGCAGAAGTGAATGAAATAGAGAATAGGTTTGATAAAACAACAAAAATGGCTGGAAAAGACTGGGTCTACGAGTTTATAAAACGACATCCTGATTTAGCTTTAAAGCAAACTACACCTACGAGCATTGCAAGAGCCATTGGTTTTAATCAAGTGCAAGTAAATAGATTTTATACAAACTTGAAAAAGTGTCAAGAAAAATATAACTTCCCTCCAGGTCGAATATATAATATGGACGAGACTGGCATCAGCACAGTGCCAAAAAAGACTCCTAAAGTTATATCACTGAAAGGAAAAAAAAATGTTAATAAAATTGTTTCTGGAGAACGTGGTCAAACTATAACTGCTGTTTGCTGTGTCAGTGCAACTGGAAATTATGTGCCTCCTCCATTTATATTTCCTCGAAAAAGGATGAAGGGTGAACTCATGGATGGAGCTCCAACTGGAAGTATTGGTATGACATCTGATTCGGGATTTATCAACACGGATTTATATTTGGAATGGCTTCATCATTTCAAGGACTACACTTCACCAACTGTCGATAATCCTGTGCTACTGATTATTGACAATCATTCAAGCCATATTAGCTTGCAAGGTACCTTGTATTGCAGACAGCATAACATCATAGTACTTACTTTGCCACCTCACAGCAGTCACAAATTACAACCCTTGGATAGGGCTATTTACAGTCCTCTTAAAAGCCAGTACGCAATTGAAGCTGATAAATGGATGGCACAGCATCCTGGTAGATCTATTAATCAATACCAAGTGACTTTGATTTTTAACAGAGCCTTTCTAAAAATTGCCACTGTGGGTAACGCAGCTTCAGCTTTCAGGGTCACGGGAATATATCCCTTCAACAACGACCTGTTTACAGAAGCAGACTTTGCACCATCTTCAGTTACAGATATGTTTTGTCCAACTTCTGATACTGTCCTTCAAAATCCGTCTACTTCAAGTTCACTTGGCTTGGATCATCATATTGTATCTGTTGCACCTGTTCCACAAGAATTAGTTGCTTCTGCGTCGGCCGATTTATCAGAAACACAAATTGAGATATCTGATGTTTCGACTGTTGCTGAAGTTCATCACCCATCAAGCTCTCCACTACCTAGAAAACCTAACATCGAAACTGATATACAGAAAGCCAATGAACCATCGCATGCGCTTTCTGATCCAATGGACCAAATAGAAGAATTTCAGGAACAACAAATTATTCCGCCTCCCGTTCTCGGTAACATTTCGAATACTCACAAATCTCATATTCCCTTAATCAATATAAGCCCATTGCCAAAGGCAACTCACATTCAGATACGACGCAAAAATGCCAGCAAATCGGAAATAATCACTAATAGTCCGTTTAAAAATATGTTAGAAGAGAAACAAAAAGAAGTCGACCGAAAATCTAAGAAAATAAGAAAACCAAAGATGAACATGGAAACCAAAACTAAGCCAAAGAAGAAAGATAATATTAAGACTAATACTAAAACTAAAGACAATGACAAGAATAAATGTTCAGAACAAGAGTACTACTGTCCGTTGTGTCAAGAAAAGTACGGAGATACGGCTGAAGCGTGGATTAAATGTTCAATTTGTAGCTCTTGGTGGCACGAAGCCTGCACCAGCTGCGAGAGTGGCATATTTACTTGTGATCTATGTGTAGTAAAAGATTGA
Protein
MPRKYIRKCPDRKVCTQEQIDKAKQLIGEGKSKRAAAEIVGINESTLRKRLKLNSTATSMGRYQQTFTREQEEEIYNHCKSSDERFYGLTLNTLRKLVYEFAEVNEIENRFDKTTKMAGKDWVYEFIKRHPDLALKQTTPTSIARAIGFNQVQVNRFYTNLKKCQEKYNFPPGRIYNMDETGISTVPKKTPKVISLKGKKNVNKIVSGERGQTITAVCCVSATGNYVPPPFIFPRKRMKGELMDGAPTGSIGMTSDSGFINTDLYLEWLHHFKDYTSPTVDNPVLLIIDNHSSHISLQGTLYCRQHNIIVLTLPPHSSHKLQPLDRAIYSPLKSQYAIEADKWMAQHPGRSINQYQVTLIFNRAFLKIATVGNAASAFRVTGIYPFNNDLFTEADFAPSSVTDMFCPTSDTVLQNPSTSSSLGLDHHIVSVAPVPQELVASASADLSETQIEISDVSTVAEVHHPSSSPLPRKPNIETDIQKANEPSHALSDPMDQIEEFQEQQIIPPPVLGNISNTHKSHIPLINISPLPKATHIQIRRKNASKSEIITNSPFKNMLEEKQKEVDRKSKKIRKPKMNMETKTKPKKKDNIKTNTKTKDNDKNKCSEQEYYCPLCQEKYGDTAEAWIKCSICSSWWHEACTSCESGIFTCDLCVVKD
Summary
Uniprot
H9JME9
H9JAE4
A0A2A4J792
J9LG75
A0A2A4J0W1
A0A2S2NVW4
+ More
A0A2H8TEI8 A0A1Y1MUL7 A0A2A4J4D1 T1EQM1 A0A1Y1M0X3 X1X8L4 J9KKA5 A0A1S3D1W0 A0A2H1VTA0 A0A1Y1LR24 A0A194RH46 A0A2H1VWF9 A0A1Y1JVK5 J9K391 A0A1B6L3T1 X1WN44 J9KHJ8 A0A1W4WFM5 T1F601 A0A0J7KEH1 X1WX11 A0A2L2YKN8 A0A2A4J1E0 A0A2A4JGA9 A0A2H1VA32 H9IUQ1 A0A3S2TQD0 A0A3S2NT42 A0A1W4WAP5 A0A2W1B5C9 A0A1W4XQ86 A0A2A4K4E0 K7JTE3 X1WM14 A0A1Y1MAV5 A0A2A4J258 A0A2A4J1M3 A0A0P6HDU5
A0A2H8TEI8 A0A1Y1MUL7 A0A2A4J4D1 T1EQM1 A0A1Y1M0X3 X1X8L4 J9KKA5 A0A1S3D1W0 A0A2H1VTA0 A0A1Y1LR24 A0A194RH46 A0A2H1VWF9 A0A1Y1JVK5 J9K391 A0A1B6L3T1 X1WN44 J9KHJ8 A0A1W4WFM5 T1F601 A0A0J7KEH1 X1WX11 A0A2L2YKN8 A0A2A4J1E0 A0A2A4JGA9 A0A2H1VA32 H9IUQ1 A0A3S2TQD0 A0A3S2NT42 A0A1W4WAP5 A0A2W1B5C9 A0A1W4XQ86 A0A2A4K4E0 K7JTE3 X1WM14 A0A1Y1MAV5 A0A2A4J258 A0A2A4J1M3 A0A0P6HDU5
EMBL
BABH01016912
BABH01009206
NWSH01002570
PCG67955.1
ABLF02009840
ABLF02016204
+ More
NWSH01004485 PCG65052.1 GGMR01008742 MBY21361.1 GFXV01000711 MBW12516.1 GEZM01026324 JAV87047.1 NWSH01003154 PCG66851.1 AMQM01000642 KB096324 ESO06516.1 GEZM01047343 JAV76877.1 ABLF02037408 ABLF02037414 ABLF02006839 ODYU01004314 SOQ44057.1 GEZM01050942 GEZM01050941 GEZM01050940 GEZM01050939 JAV75288.1 KQ460205 KPJ16902.1 ODYU01004859 SOQ45169.1 GEZM01099403 GEZM01099402 JAV53344.1 ABLF02011245 GEBQ01021595 JAT18382.1 ABLF02017996 ABLF02011868 ABLF02011869 AMQM01004377 KB096502 ESO04455.1 LBMM01008804 KMQ88629.1 ABLF02062637 IAAA01031058 LAA08688.1 NWSH01003711 PCG65895.1 NWSH01001638 PCG70614.1 ODYU01001473 SOQ37710.1 BABH01000686 RSAL01000024 RVE52221.1 RSAL01000230 RVE43880.1 KZ150321 PZC71419.1 NWSH01000193 PCG78522.1 ABLF02007358 ABLF02007359 ABLF02007364 ABLF02037183 GEZM01039017 JAV81445.1 NWSH01003844 PCG65768.1 PCG65769.1 GDIQ01020929 JAN73808.1
NWSH01004485 PCG65052.1 GGMR01008742 MBY21361.1 GFXV01000711 MBW12516.1 GEZM01026324 JAV87047.1 NWSH01003154 PCG66851.1 AMQM01000642 KB096324 ESO06516.1 GEZM01047343 JAV76877.1 ABLF02037408 ABLF02037414 ABLF02006839 ODYU01004314 SOQ44057.1 GEZM01050942 GEZM01050941 GEZM01050940 GEZM01050939 JAV75288.1 KQ460205 KPJ16902.1 ODYU01004859 SOQ45169.1 GEZM01099403 GEZM01099402 JAV53344.1 ABLF02011245 GEBQ01021595 JAT18382.1 ABLF02017996 ABLF02011868 ABLF02011869 AMQM01004377 KB096502 ESO04455.1 LBMM01008804 KMQ88629.1 ABLF02062637 IAAA01031058 LAA08688.1 NWSH01003711 PCG65895.1 NWSH01001638 PCG70614.1 ODYU01001473 SOQ37710.1 BABH01000686 RSAL01000024 RVE52221.1 RSAL01000230 RVE43880.1 KZ150321 PZC71419.1 NWSH01000193 PCG78522.1 ABLF02007358 ABLF02007359 ABLF02007364 ABLF02037183 GEZM01039017 JAV81445.1 NWSH01003844 PCG65768.1 PCG65769.1 GDIQ01020929 JAN73808.1
Proteomes
Interpro
IPR004875
DDE_SF_endonuclease_dom
+ More
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR013083 Znf_RING/FYVE/PHD
IPR006600 HTH_CenpB_DNA-bd_dom
IPR036880 Kunitz_BPTI_sf
IPR001965 Znf_PHD
IPR007110 Ig-like_dom
IPR019787 Znf_PHD-finger
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR013083 Znf_RING/FYVE/PHD
IPR006600 HTH_CenpB_DNA-bd_dom
IPR036880 Kunitz_BPTI_sf
IPR001965 Znf_PHD
IPR007110 Ig-like_dom
IPR019787 Znf_PHD-finger
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
Gene 3D
ProteinModelPortal
H9JME9
H9JAE4
A0A2A4J792
J9LG75
A0A2A4J0W1
A0A2S2NVW4
+ More
A0A2H8TEI8 A0A1Y1MUL7 A0A2A4J4D1 T1EQM1 A0A1Y1M0X3 X1X8L4 J9KKA5 A0A1S3D1W0 A0A2H1VTA0 A0A1Y1LR24 A0A194RH46 A0A2H1VWF9 A0A1Y1JVK5 J9K391 A0A1B6L3T1 X1WN44 J9KHJ8 A0A1W4WFM5 T1F601 A0A0J7KEH1 X1WX11 A0A2L2YKN8 A0A2A4J1E0 A0A2A4JGA9 A0A2H1VA32 H9IUQ1 A0A3S2TQD0 A0A3S2NT42 A0A1W4WAP5 A0A2W1B5C9 A0A1W4XQ86 A0A2A4K4E0 K7JTE3 X1WM14 A0A1Y1MAV5 A0A2A4J258 A0A2A4J1M3 A0A0P6HDU5
A0A2H8TEI8 A0A1Y1MUL7 A0A2A4J4D1 T1EQM1 A0A1Y1M0X3 X1X8L4 J9KKA5 A0A1S3D1W0 A0A2H1VTA0 A0A1Y1LR24 A0A194RH46 A0A2H1VWF9 A0A1Y1JVK5 J9K391 A0A1B6L3T1 X1WN44 J9KHJ8 A0A1W4WFM5 T1F601 A0A0J7KEH1 X1WX11 A0A2L2YKN8 A0A2A4J1E0 A0A2A4JGA9 A0A2H1VA32 H9IUQ1 A0A3S2TQD0 A0A3S2NT42 A0A1W4WAP5 A0A2W1B5C9 A0A1W4XQ86 A0A2A4K4E0 K7JTE3 X1WM14 A0A1Y1MAV5 A0A2A4J258 A0A2A4J1M3 A0A0P6HDU5
Ontologies
GO
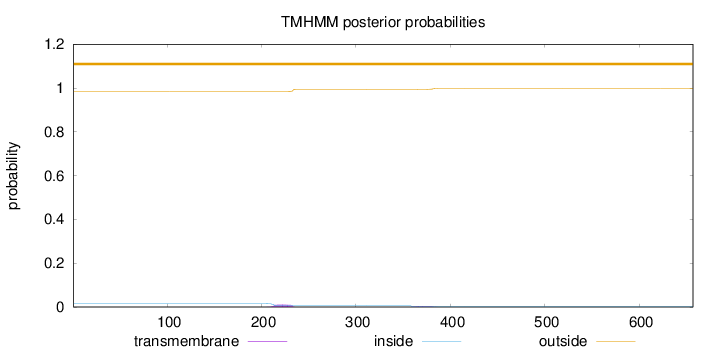
Topology
Subcellular location
Nucleus
Length:
657
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31548
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01574
outside
1 - 657
Population Genetic Test Statistics
Pi
58.352671
Theta
157.809269
Tajima's D
-1.131979
CLR
0
CSRT
0.112944352782361
Interpretation
Uncertain
