Gene
KWMTBOMO15634
Pre Gene Modal
BGIBMGA010703
Annotation
PREDICTED:_mpv17-like_protein_2_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.81
Sequence
CDS
ATGTCGGGGATTCTCTCGAAAGTTTTGAGGTTACCACGGAAATATCCTTTGCTTCGCGGTATGCTGTGTTACGCGGTGATATGGCCCGCTTGCAGCATAACCCAAGAGTTCATGGAGCACGGACCCAACTTGGAAAAATTGGATTGGGCTAGAGCAACGCGCTTTGGCGTGTTTGGAACATTCTTTATGGCTCCAGTCTTTTACGGGTGGATGAAGTATAGCAGCAGATTTTTCAAGAGAAATAATTTGAAGACAGCAATCCTCAGGGCGGGCTTAGAACAAATCTCTTACGCTCCTCTAGCGACGGCGTATTTCTTCTTCGGCATGAGTCTGCTCGAGATGAAGCCTATAGAAGCATGCCTAAAAGAAGTGGAAGAAAAACTCTGGCCCACGTACAAAATAGGGGTGTTATTCTGGCCGATGGCTCAAACTGTTAACTTTTATTTTGTTTCCGAAAAGAACAGGATAGTATTTGTGAGCTTGGCTAGTTTTGTATGGACGGTTTATCTCGCGCATGTTAAAAGTAAAGAAAAAAATGAAAAAGCTGTAATTTGA
Protein
MSGILSKVLRLPRKYPLLRGMLCYAVIWPACSITQEFMEHGPNLEKLDWARATRFGVFGTFFMAPVFYGWMKYSSRFFKRNNLKTAILRAGLEQISYAPLATAYFFFGMSLLEMKPIEACLKEVEEKLWPTYKIGVLFWPMAQTVNFYFVSEKNRIVFVSLASFVWTVYLAHVKSKEKNEKAVI
Summary
Similarity
Belongs to the peroxisomal membrane protein PXMP2/4 family.
Uniprot
H9JMF0
A0A2A4JGU9
A0A0N1IJD6
A0A0L7KX04
A0A194QCH4
A0A212EW96
+ More
K7IVG5 A0A151XD96 A0A088AQA7 A0A0L7QZ09 A0A0K8TQ11 A0A0M8ZUF4 Q9XZT2 A0A3B0KQX1 F4WSR8 A0A1I8MN04 Q9W588 A0A1W4VFM6 A0A232EZ88 B3MRG6 A0A151J163 A0A0P8XK46 E9IJN4 Q29J07 B4I9E7 B4GTK9 D6X0W2 A0A195D2Q0 A0A158NHA5 A0A0Q9XF68 B4PXP9 B3P9I5 B4L755 A0A0Q9XHR7 A0A1I8P3M2 A0A026X4T6 A0A3L8DTM0 A0A067QT30 A0A1B0BZ07 B4MB27 A0A0A9X194 A0A0C9RNK6 A0A1A9ULM9 B4JLQ6 A0A1B0FAJ2 A0A1I8PYC4 A0A1Q3F7V2 B4NCJ9 A0A2A3EI42 T1E391 A0A1I8PYF2 Q17KN8 A0A182FN37 A0A0K8VT58 A0A1Y1KE00 R4FL62 A0A0P4WFS6 A0A0P4WAE6 A0A034WGR2 A0A1I8JU85 A0A1A9ULM6 A0A1Y1KHG1 A0A0A1X3L9 A0A1B0FAJ5 A0A1A9X1D3 A0A182NNS4 A0A182QFQ3 A0A1A9YQ86 A0A1B0DNN7 A0A1B0A0A3 A0A154NY34 A0A1W4XSZ1 A0A182XH92 A0A182HR14 Q7PDE1 A0A182PVP8 A0A2J7R306 A0A1W7R4V8 A0A034VMM7 A0A3S2LES6 A0A0J7L3C6 A0A1Y9IW50 W8C6Y9 E2BJR8 A0A212EUA2 A0A1B6GFQ9 A0A2H1VH53 A0A1I8M4T7 A0A0M4F8D0 A0A1B6KRL9 A0A0P5EAY4 A0A1I8PQK7 A0A1B0BZ08 A0A0P5FLV0 A0A3B0KDF7 W8BK13 Q29J09 B3MRG4 A0A2R7W292 B3P9I7
K7IVG5 A0A151XD96 A0A088AQA7 A0A0L7QZ09 A0A0K8TQ11 A0A0M8ZUF4 Q9XZT2 A0A3B0KQX1 F4WSR8 A0A1I8MN04 Q9W588 A0A1W4VFM6 A0A232EZ88 B3MRG6 A0A151J163 A0A0P8XK46 E9IJN4 Q29J07 B4I9E7 B4GTK9 D6X0W2 A0A195D2Q0 A0A158NHA5 A0A0Q9XF68 B4PXP9 B3P9I5 B4L755 A0A0Q9XHR7 A0A1I8P3M2 A0A026X4T6 A0A3L8DTM0 A0A067QT30 A0A1B0BZ07 B4MB27 A0A0A9X194 A0A0C9RNK6 A0A1A9ULM9 B4JLQ6 A0A1B0FAJ2 A0A1I8PYC4 A0A1Q3F7V2 B4NCJ9 A0A2A3EI42 T1E391 A0A1I8PYF2 Q17KN8 A0A182FN37 A0A0K8VT58 A0A1Y1KE00 R4FL62 A0A0P4WFS6 A0A0P4WAE6 A0A034WGR2 A0A1I8JU85 A0A1A9ULM6 A0A1Y1KHG1 A0A0A1X3L9 A0A1B0FAJ5 A0A1A9X1D3 A0A182NNS4 A0A182QFQ3 A0A1A9YQ86 A0A1B0DNN7 A0A1B0A0A3 A0A154NY34 A0A1W4XSZ1 A0A182XH92 A0A182HR14 Q7PDE1 A0A182PVP8 A0A2J7R306 A0A1W7R4V8 A0A034VMM7 A0A3S2LES6 A0A0J7L3C6 A0A1Y9IW50 W8C6Y9 E2BJR8 A0A212EUA2 A0A1B6GFQ9 A0A2H1VH53 A0A1I8M4T7 A0A0M4F8D0 A0A1B6KRL9 A0A0P5EAY4 A0A1I8PQK7 A0A1B0BZ08 A0A0P5FLV0 A0A3B0KDF7 W8BK13 Q29J09 B3MRG4 A0A2R7W292 B3P9I7
Pubmed
19121390
26354079
26227816
22118469
20075255
26369729
+ More
21719571 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 17994087 21282665 15632085 23185243 18362917 19820115 21347285 17550304 18057021 24508170 30249741 24845553 25401762 26823975 24330624 17510324 28004739 25348373 25830018 12364791 14747013 17210077 24495485 20798317
21719571 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 17994087 21282665 15632085 23185243 18362917 19820115 21347285 17550304 18057021 24508170 30249741 24845553 25401762 26823975 24330624 17510324 28004739 25348373 25830018 12364791 14747013 17210077 24495485 20798317
EMBL
BABH01016908
NWSH01001415
PCG71305.1
KQ459941
KPJ19156.1
JTDY01004929
+ More
KOB67584.1 KQ459193 KPJ03167.1 AGBW02012063 OWR45741.1 KQ982294 KYQ58341.1 KQ414685 KOC63833.1 GDAI01001149 JAI16454.1 KQ435840 KOX71365.1 AL031027 CAB41536.1 OUUW01000011 SPP86288.1 GL888327 EGI62725.1 AE014298 BT003441 BT025917 AAF45630.1 AAO39444.1 ABG02161.1 ACZ95171.1 NNAY01001524 OXU23691.1 CH902622 EDV34371.1 KQ980555 KYN15560.1 KPU75103.1 GL763877 EFZ19225.1 CH379063 EAL32494.2 KRT06090.1 CH480825 EDW43828.1 CH479190 EDW25879.1 KQ971372 EFA09541.1 KQ976973 KYN06644.1 ADTU01015421 CH933812 KRG07179.1 CM000162 EDX00902.1 KRK05811.1 KRK05812.1 CH954183 EDV45481.1 KQS25975.1 KQS25976.1 EDW06201.2 KRG07178.1 KRG07180.1 KRG07177.1 KK107020 EZA62439.1 QOIP01000004 RLU23249.1 KK853051 KDR12034.1 JXJN01022928 CH940655 EDW66436.1 KRF82677.1 GBHO01041013 GBHO01032731 GBHO01032730 GBHO01032729 GBHO01032727 GBHO01021525 GBHO01021524 GBHO01021523 GBHO01021522 GBHO01021521 GBRD01016626 GBRD01016624 GDHC01008942 JAG02591.1 JAG10873.1 JAG10874.1 JAG10875.1 JAG10877.1 JAG22079.1 JAG22080.1 JAG22081.1 JAG22082.1 JAG22083.1 JAG49200.1 JAQ09687.1 GBYB01009985 JAG79752.1 CH916371 EDV91667.1 CCAG010006944 GFDL01011428 JAV23617.1 CH964239 EDW82558.1 KZ288232 PBC31455.1 GALA01000054 JAA94798.1 CH477222 EAT47266.1 GDHF01032930 GDHF01010250 JAI19384.1 JAI42064.1 GEZM01086214 JAV59669.1 ACPB03000425 GAHY01002224 JAA75286.1 GDRN01058785 JAI65561.1 GDRN01058784 JAI65562.1 GAKP01004216 GAKP01004215 JAC54736.1 GEZM01086213 JAV59670.1 GBXI01008811 JAD05481.1 AXCN02001979 AJVK01001555 KQ434782 KZC04579.1 APCN01001632 AAAB01008980 EAA13780.4 NEVH01007824 PNF35205.1 GEHC01001448 JAV46197.1 GAKP01014368 GAKP01014367 GAKP01014366 JAC44586.1 RSAL01000172 RVE45139.1 LBMM01000855 KMQ97372.1 GAMC01000847 JAC05709.1 GL448636 EFN84043.1 AGBW02012455 OWR45031.1 GECZ01008518 JAS61251.1 ODYU01002544 SOQ40180.1 CP012528 ALC48245.1 GEBQ01025900 JAT14077.1 GDIP01144062 GDIQ01037463 JAJ79340.1 JAN57274.1 GDIQ01255112 JAJ96612.1 SPP86290.1 GAMC01007503 GAMC01007502 JAB99052.1 EAL32493.1 EDV34369.1 KK854259 PTY13853.1 EDV45483.1
KOB67584.1 KQ459193 KPJ03167.1 AGBW02012063 OWR45741.1 KQ982294 KYQ58341.1 KQ414685 KOC63833.1 GDAI01001149 JAI16454.1 KQ435840 KOX71365.1 AL031027 CAB41536.1 OUUW01000011 SPP86288.1 GL888327 EGI62725.1 AE014298 BT003441 BT025917 AAF45630.1 AAO39444.1 ABG02161.1 ACZ95171.1 NNAY01001524 OXU23691.1 CH902622 EDV34371.1 KQ980555 KYN15560.1 KPU75103.1 GL763877 EFZ19225.1 CH379063 EAL32494.2 KRT06090.1 CH480825 EDW43828.1 CH479190 EDW25879.1 KQ971372 EFA09541.1 KQ976973 KYN06644.1 ADTU01015421 CH933812 KRG07179.1 CM000162 EDX00902.1 KRK05811.1 KRK05812.1 CH954183 EDV45481.1 KQS25975.1 KQS25976.1 EDW06201.2 KRG07178.1 KRG07180.1 KRG07177.1 KK107020 EZA62439.1 QOIP01000004 RLU23249.1 KK853051 KDR12034.1 JXJN01022928 CH940655 EDW66436.1 KRF82677.1 GBHO01041013 GBHO01032731 GBHO01032730 GBHO01032729 GBHO01032727 GBHO01021525 GBHO01021524 GBHO01021523 GBHO01021522 GBHO01021521 GBRD01016626 GBRD01016624 GDHC01008942 JAG02591.1 JAG10873.1 JAG10874.1 JAG10875.1 JAG10877.1 JAG22079.1 JAG22080.1 JAG22081.1 JAG22082.1 JAG22083.1 JAG49200.1 JAQ09687.1 GBYB01009985 JAG79752.1 CH916371 EDV91667.1 CCAG010006944 GFDL01011428 JAV23617.1 CH964239 EDW82558.1 KZ288232 PBC31455.1 GALA01000054 JAA94798.1 CH477222 EAT47266.1 GDHF01032930 GDHF01010250 JAI19384.1 JAI42064.1 GEZM01086214 JAV59669.1 ACPB03000425 GAHY01002224 JAA75286.1 GDRN01058785 JAI65561.1 GDRN01058784 JAI65562.1 GAKP01004216 GAKP01004215 JAC54736.1 GEZM01086213 JAV59670.1 GBXI01008811 JAD05481.1 AXCN02001979 AJVK01001555 KQ434782 KZC04579.1 APCN01001632 AAAB01008980 EAA13780.4 NEVH01007824 PNF35205.1 GEHC01001448 JAV46197.1 GAKP01014368 GAKP01014367 GAKP01014366 JAC44586.1 RSAL01000172 RVE45139.1 LBMM01000855 KMQ97372.1 GAMC01000847 JAC05709.1 GL448636 EFN84043.1 AGBW02012455 OWR45031.1 GECZ01008518 JAS61251.1 ODYU01002544 SOQ40180.1 CP012528 ALC48245.1 GEBQ01025900 JAT14077.1 GDIP01144062 GDIQ01037463 JAJ79340.1 JAN57274.1 GDIQ01255112 JAJ96612.1 SPP86290.1 GAMC01007503 GAMC01007502 JAB99052.1 EAL32493.1 EDV34369.1 KK854259 PTY13853.1 EDV45483.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000053268
UP000007151
+ More
UP000002358 UP000075809 UP000005203 UP000053825 UP000053105 UP000268350 UP000007755 UP000095301 UP000000803 UP000192221 UP000215335 UP000007801 UP000078492 UP000001819 UP000001292 UP000008744 UP000007266 UP000078542 UP000005205 UP000009192 UP000002282 UP000008711 UP000095300 UP000053097 UP000279307 UP000027135 UP000092460 UP000008792 UP000078200 UP000001070 UP000092444 UP000007798 UP000242457 UP000008820 UP000069272 UP000015103 UP000075900 UP000091820 UP000075884 UP000075886 UP000092443 UP000092462 UP000092445 UP000076502 UP000192223 UP000076407 UP000075840 UP000007062 UP000075885 UP000235965 UP000283053 UP000036403 UP000075920 UP000008237 UP000092553
UP000002358 UP000075809 UP000005203 UP000053825 UP000053105 UP000268350 UP000007755 UP000095301 UP000000803 UP000192221 UP000215335 UP000007801 UP000078492 UP000001819 UP000001292 UP000008744 UP000007266 UP000078542 UP000005205 UP000009192 UP000002282 UP000008711 UP000095300 UP000053097 UP000279307 UP000027135 UP000092460 UP000008792 UP000078200 UP000001070 UP000092444 UP000007798 UP000242457 UP000008820 UP000069272 UP000015103 UP000075900 UP000091820 UP000075884 UP000075886 UP000092443 UP000092462 UP000092445 UP000076502 UP000192223 UP000076407 UP000075840 UP000007062 UP000075885 UP000235965 UP000283053 UP000036403 UP000075920 UP000008237 UP000092553
Pfam
PF04117 Mpv17_PMP22
SUPFAM
SSF46934
SSF46934
ProteinModelPortal
H9JMF0
A0A2A4JGU9
A0A0N1IJD6
A0A0L7KX04
A0A194QCH4
A0A212EW96
+ More
K7IVG5 A0A151XD96 A0A088AQA7 A0A0L7QZ09 A0A0K8TQ11 A0A0M8ZUF4 Q9XZT2 A0A3B0KQX1 F4WSR8 A0A1I8MN04 Q9W588 A0A1W4VFM6 A0A232EZ88 B3MRG6 A0A151J163 A0A0P8XK46 E9IJN4 Q29J07 B4I9E7 B4GTK9 D6X0W2 A0A195D2Q0 A0A158NHA5 A0A0Q9XF68 B4PXP9 B3P9I5 B4L755 A0A0Q9XHR7 A0A1I8P3M2 A0A026X4T6 A0A3L8DTM0 A0A067QT30 A0A1B0BZ07 B4MB27 A0A0A9X194 A0A0C9RNK6 A0A1A9ULM9 B4JLQ6 A0A1B0FAJ2 A0A1I8PYC4 A0A1Q3F7V2 B4NCJ9 A0A2A3EI42 T1E391 A0A1I8PYF2 Q17KN8 A0A182FN37 A0A0K8VT58 A0A1Y1KE00 R4FL62 A0A0P4WFS6 A0A0P4WAE6 A0A034WGR2 A0A1I8JU85 A0A1A9ULM6 A0A1Y1KHG1 A0A0A1X3L9 A0A1B0FAJ5 A0A1A9X1D3 A0A182NNS4 A0A182QFQ3 A0A1A9YQ86 A0A1B0DNN7 A0A1B0A0A3 A0A154NY34 A0A1W4XSZ1 A0A182XH92 A0A182HR14 Q7PDE1 A0A182PVP8 A0A2J7R306 A0A1W7R4V8 A0A034VMM7 A0A3S2LES6 A0A0J7L3C6 A0A1Y9IW50 W8C6Y9 E2BJR8 A0A212EUA2 A0A1B6GFQ9 A0A2H1VH53 A0A1I8M4T7 A0A0M4F8D0 A0A1B6KRL9 A0A0P5EAY4 A0A1I8PQK7 A0A1B0BZ08 A0A0P5FLV0 A0A3B0KDF7 W8BK13 Q29J09 B3MRG4 A0A2R7W292 B3P9I7
K7IVG5 A0A151XD96 A0A088AQA7 A0A0L7QZ09 A0A0K8TQ11 A0A0M8ZUF4 Q9XZT2 A0A3B0KQX1 F4WSR8 A0A1I8MN04 Q9W588 A0A1W4VFM6 A0A232EZ88 B3MRG6 A0A151J163 A0A0P8XK46 E9IJN4 Q29J07 B4I9E7 B4GTK9 D6X0W2 A0A195D2Q0 A0A158NHA5 A0A0Q9XF68 B4PXP9 B3P9I5 B4L755 A0A0Q9XHR7 A0A1I8P3M2 A0A026X4T6 A0A3L8DTM0 A0A067QT30 A0A1B0BZ07 B4MB27 A0A0A9X194 A0A0C9RNK6 A0A1A9ULM9 B4JLQ6 A0A1B0FAJ2 A0A1I8PYC4 A0A1Q3F7V2 B4NCJ9 A0A2A3EI42 T1E391 A0A1I8PYF2 Q17KN8 A0A182FN37 A0A0K8VT58 A0A1Y1KE00 R4FL62 A0A0P4WFS6 A0A0P4WAE6 A0A034WGR2 A0A1I8JU85 A0A1A9ULM6 A0A1Y1KHG1 A0A0A1X3L9 A0A1B0FAJ5 A0A1A9X1D3 A0A182NNS4 A0A182QFQ3 A0A1A9YQ86 A0A1B0DNN7 A0A1B0A0A3 A0A154NY34 A0A1W4XSZ1 A0A182XH92 A0A182HR14 Q7PDE1 A0A182PVP8 A0A2J7R306 A0A1W7R4V8 A0A034VMM7 A0A3S2LES6 A0A0J7L3C6 A0A1Y9IW50 W8C6Y9 E2BJR8 A0A212EUA2 A0A1B6GFQ9 A0A2H1VH53 A0A1I8M4T7 A0A0M4F8D0 A0A1B6KRL9 A0A0P5EAY4 A0A1I8PQK7 A0A1B0BZ08 A0A0P5FLV0 A0A3B0KDF7 W8BK13 Q29J09 B3MRG4 A0A2R7W292 B3P9I7
Ontologies
GO
PANTHER
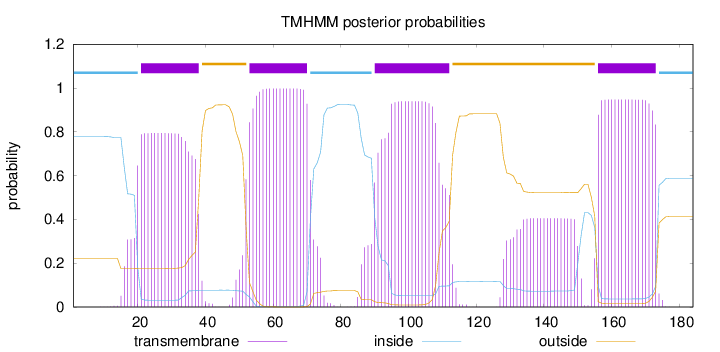
Topology
Length:
184
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
82.027
Exp number, first 60 AAs:
24.39845
Total prob of N-in:
0.77870
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 38
outside
39 - 52
TMhelix
53 - 70
inside
71 - 89
TMhelix
90 - 112
outside
113 - 155
TMhelix
156 - 173
inside
174 - 184
Population Genetic Test Statistics
Pi
152.190128
Theta
186.575435
Tajima's D
-0.736121
CLR
163.965547
CSRT
0.191540422978851
Interpretation
Uncertain
