Gene
KWMTBOMO15630
Pre Gene Modal
BGIBMGA010707
Annotation
PREDICTED:_2-aminoethanethiol_dioxygenase_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.089
Sequence
CDS
ATGGATATTAAGAATGAAACCAAGCCAACCATGGAAATTACTAACATACCGCCTATTGTATCAATATACCGGCACGCAATGCGCACCTTTGATGACAATTTCAAAATGGATTTCACGACCAACTTAAATAAGTTAAAATCTATGATGGATCACCTCAAAGCTGAAGATTTCGCATTCGATAATAGCCTCAACGATATCGAGACATGGAGGCAGCCGCTTAAAGCCCCATGTACGTACATTAAAGTGTTTGAAGACTATCAGATAAATATGAGCATTTTCGTGTTAAAACCTGGTTTTCGAATGCCTCTCCACGATCACCCTCGTATGCATGGACTATTGAAAGTGATTACTGGTGCAGTGAATATACGGAGTTTCTCTGAGTACCCATTAACAGAACAGGTGAGTAGCTTAGACCTCAAGGTGCGCGCGAGATTCGAGGCAGCTCGTTTGGCACGGGGTATTCACAGAAGGAGAAGGCTTTTCGCGGAAGTGACGGCTGATCGTGTGTGTAAGGCCAATGCTGAGACCTGCATCCTCACTCCAACAATATCAAATTACCACGAAATCGAGGCCTTGGATATGCCTGCAGCATTCTTTGATATACTTTCACCACCATACGATACGATGATTGAAGGTATAGGTCCTAGAAGGTGCCGGTATTATGAAGTGGCTAACGTAATAAGTAGTAATCTAGTTGAATTGGAGGAGATAGAAGTGCCAAAATGTTTTTATTGTGATCAAGCACCATACTTAGGGCCCACACTTGCTTAG
Protein
MDIKNETKPTMEITNIPPIVSIYRHAMRTFDDNFKMDFTTNLNKLKSMMDHLKAEDFAFDNSLNDIETWRQPLKAPCTYIKVFEDYQINMSIFVLKPGFRMPLHDHPRMHGLLKVITGAVNIRSFSEYPLTEQVSSLDLKVRARFEAARLARGIHRRRRLFAEVTADRVCKANAETCILTPTISNYHEIEALDMPAAFFDILSPPYDTMIEGIGPRRCRYYEVANVISSNLVELEEIEVPKCFYCDQAPYLGPTLA
Summary
Uniprot
H9JMF4
A0A2A4ITF7
A0A2H1VNS3
A0A0N0PEH3
A0A1E1WJG5
A0A194QCL8
+ More
A0A0L7LQX3 A0A1E1W058 A0A212EWC3 A0A1W4WVA0 V5GK94 A0A1Y1JVC2 D6WZW8 Q17FN6 Q16WG8 A0A0K8TL44 A0A0P6IUE8 A0A182GF60 A0A1A9VN32 A0A1B0G3Y8 A0A1A9ZMJ9 A0A0L0C202 A0A1Q3F5R5 N6U292 A0A1B0AQB1 A0A0K8UZC4 A0A1I8MF42 A0A182PIV8 A0A182X277 B0WC79 A0A182U4Z3 A0A182V7B9 A0A182K0P8 A0NBN0 A0A182I6D7 A0A034WKT2 A0A3F2YQA3 E2BKW6 A0A182Y886 A0A1B6L6N8 A0A182R6P9 A0A084WMV9 A0A2M4AZ60 A0A0J7N0L6 A0A182WI33 A0A182SYC0 A0A182MEH2 A0A2M4CP26 A0A1B6GJV6 W5JL03 A0A2M4CNG6 A0A2M4CPI7 A0A1B6JJP4 A0A182JB67 V4BNI4 E2ACF2 A0A182N7F7 A0A182QAW1 A0A0P4VJQ5 R4G7Y4 A0A2P6KAV8 B4MXE0 A0A1B0CQG2 A0A1L8DB51 A0A023F8R6 A0A2S2QMK6 T1DEZ6 A0A1B6CV59 A0A067QV27 A0A232F4D4 A0A0L8HRA0 K7J5B6 A0A195E3R6 R7TVH5 A0A0M4E8X3 A0A158NW35 A0A2R5LIN1 A0A1I8PZM3 A0A195CS53 B4LGZ5 A0A151WVK3 U5EWC7 A0A151I0T0 A0A2J7QKL0 T1KXA8 A0A2A3EB42 A0A1Z5LE34 A0A2R7X2P3 B4IWR4 W4YHW1 A0A2P8Z212 A0A1W4VNG9 A0A2S2RA44 B4KWX7 A0A151JYD8 A0A0L7QUU3 A0A1E1XQ38 A0A3R7QAD7
A0A0L7LQX3 A0A1E1W058 A0A212EWC3 A0A1W4WVA0 V5GK94 A0A1Y1JVC2 D6WZW8 Q17FN6 Q16WG8 A0A0K8TL44 A0A0P6IUE8 A0A182GF60 A0A1A9VN32 A0A1B0G3Y8 A0A1A9ZMJ9 A0A0L0C202 A0A1Q3F5R5 N6U292 A0A1B0AQB1 A0A0K8UZC4 A0A1I8MF42 A0A182PIV8 A0A182X277 B0WC79 A0A182U4Z3 A0A182V7B9 A0A182K0P8 A0NBN0 A0A182I6D7 A0A034WKT2 A0A3F2YQA3 E2BKW6 A0A182Y886 A0A1B6L6N8 A0A182R6P9 A0A084WMV9 A0A2M4AZ60 A0A0J7N0L6 A0A182WI33 A0A182SYC0 A0A182MEH2 A0A2M4CP26 A0A1B6GJV6 W5JL03 A0A2M4CNG6 A0A2M4CPI7 A0A1B6JJP4 A0A182JB67 V4BNI4 E2ACF2 A0A182N7F7 A0A182QAW1 A0A0P4VJQ5 R4G7Y4 A0A2P6KAV8 B4MXE0 A0A1B0CQG2 A0A1L8DB51 A0A023F8R6 A0A2S2QMK6 T1DEZ6 A0A1B6CV59 A0A067QV27 A0A232F4D4 A0A0L8HRA0 K7J5B6 A0A195E3R6 R7TVH5 A0A0M4E8X3 A0A158NW35 A0A2R5LIN1 A0A1I8PZM3 A0A195CS53 B4LGZ5 A0A151WVK3 U5EWC7 A0A151I0T0 A0A2J7QKL0 T1KXA8 A0A2A3EB42 A0A1Z5LE34 A0A2R7X2P3 B4IWR4 W4YHW1 A0A2P8Z212 A0A1W4VNG9 A0A2S2RA44 B4KWX7 A0A151JYD8 A0A0L7QUU3 A0A1E1XQ38 A0A3R7QAD7
Pubmed
19121390
26354079
26227816
22118469
28004739
18362917
+ More
19820115 17510324 26369729 26999592 26483478 26108605 23537049 25315136 12364791 14747013 17210077 25348373 20798317 25244985 24438588 20920257 23761445 23254933 27129103 17994087 25474469 24845553 28648823 20075255 21347285 28528879 29403074 29209593
19820115 17510324 26369729 26999592 26483478 26108605 23537049 25315136 12364791 14747013 17210077 25348373 20798317 25244985 24438588 20920257 23761445 23254933 27129103 17994087 25474469 24845553 28648823 20075255 21347285 28528879 29403074 29209593
EMBL
BABH01016898
NWSH01007463
PCG62959.1
ODYU01003314
SOQ41904.1
KQ459941
+ More
KPJ19153.1 GDQN01004018 JAT87036.1 KQ459193 KPJ03164.1 JTDY01000287 KOB77898.1 GDQN01010730 JAT80324.1 AGBW02012063 OWR45744.1 GALX01006444 JAB62022.1 GEZM01102898 JAV51740.1 KQ971372 EFA09634.1 CH477269 EAT45388.1 CH477566 EAT38929.1 GDAI01002710 JAI14893.1 GDUN01000465 JAN95454.1 JXUM01059290 KQ562045 KXJ76818.1 CCAG010017310 JRES01000997 KNC26291.1 GFDL01012176 JAV22869.1 APGK01052696 KB741213 KB631874 ENN72672.1 ERL86867.1 JXJN01001782 GDHF01020421 JAI31893.1 DS231886 EDS43324.1 AAAB01008807 EAU77700.2 APCN01003620 GAKP01003703 JAC55249.1 GL448836 EFN83662.1 GEBQ01020589 GEBQ01003154 JAT19388.1 JAT36823.1 ATLV01024521 KE525352 KFB51553.1 GGFK01012537 MBW45858.1 LBMM01012507 KMQ86185.1 AXCM01002040 GGFL01002896 MBW67074.1 GECZ01007033 JAS62736.1 ADMH02001223 ETN63595.1 GGFL01002641 MBW66819.1 GGFL01002640 MBW66818.1 GECU01008296 JAS99410.1 KB202444 ESO90404.1 GL438494 EFN68891.1 AXCN02000573 GDKW01002133 JAI54462.1 ACPB03007651 GAHY01001917 JAA75593.1 MWRG01017583 PRD23474.1 CH963876 EDW76709.1 AJWK01023506 GFDF01010398 JAV03686.1 GBBI01000927 JAC17785.1 GGMS01009169 MBY78372.1 GAMD01003316 JAA98274.1 GEDC01026311 GEDC01022393 GEDC01019938 GEDC01010939 JAS10987.1 JAS14905.1 JAS17360.1 JAS26359.1 KK852902 KDR14049.1 NNAY01001051 OXU25330.1 KQ417492 KOF91717.1 AAZX01012106 KQ979701 KYN19514.1 AMQN01010754 KB308480 ELT97719.1 CP012525 ALC43428.1 ADTU01027806 GGLE01005061 MBY09187.1 KQ977329 KYN03526.1 CH940647 EDW69485.1 KQ982718 KYQ51705.1 GANO01000627 JAB59244.1 KQ976606 KYM79325.1 NEVH01013262 PNF29124.1 CAEY01000677 KZ288293 PBC28993.1 GFJQ02001298 JAW05672.1 KK856508 PTY25931.1 CH916366 EDV96290.1 AAGJ04064386 PYGN01000235 PSN50528.1 GGMS01017019 MBY86222.1 CH933809 EDW18598.1 KQ981477 KYN41442.1 KQ414731 KOC62427.1 GFAA01002002 JAU01433.1 QCYY01002113 ROT72779.1
KPJ19153.1 GDQN01004018 JAT87036.1 KQ459193 KPJ03164.1 JTDY01000287 KOB77898.1 GDQN01010730 JAT80324.1 AGBW02012063 OWR45744.1 GALX01006444 JAB62022.1 GEZM01102898 JAV51740.1 KQ971372 EFA09634.1 CH477269 EAT45388.1 CH477566 EAT38929.1 GDAI01002710 JAI14893.1 GDUN01000465 JAN95454.1 JXUM01059290 KQ562045 KXJ76818.1 CCAG010017310 JRES01000997 KNC26291.1 GFDL01012176 JAV22869.1 APGK01052696 KB741213 KB631874 ENN72672.1 ERL86867.1 JXJN01001782 GDHF01020421 JAI31893.1 DS231886 EDS43324.1 AAAB01008807 EAU77700.2 APCN01003620 GAKP01003703 JAC55249.1 GL448836 EFN83662.1 GEBQ01020589 GEBQ01003154 JAT19388.1 JAT36823.1 ATLV01024521 KE525352 KFB51553.1 GGFK01012537 MBW45858.1 LBMM01012507 KMQ86185.1 AXCM01002040 GGFL01002896 MBW67074.1 GECZ01007033 JAS62736.1 ADMH02001223 ETN63595.1 GGFL01002641 MBW66819.1 GGFL01002640 MBW66818.1 GECU01008296 JAS99410.1 KB202444 ESO90404.1 GL438494 EFN68891.1 AXCN02000573 GDKW01002133 JAI54462.1 ACPB03007651 GAHY01001917 JAA75593.1 MWRG01017583 PRD23474.1 CH963876 EDW76709.1 AJWK01023506 GFDF01010398 JAV03686.1 GBBI01000927 JAC17785.1 GGMS01009169 MBY78372.1 GAMD01003316 JAA98274.1 GEDC01026311 GEDC01022393 GEDC01019938 GEDC01010939 JAS10987.1 JAS14905.1 JAS17360.1 JAS26359.1 KK852902 KDR14049.1 NNAY01001051 OXU25330.1 KQ417492 KOF91717.1 AAZX01012106 KQ979701 KYN19514.1 AMQN01010754 KB308480 ELT97719.1 CP012525 ALC43428.1 ADTU01027806 GGLE01005061 MBY09187.1 KQ977329 KYN03526.1 CH940647 EDW69485.1 KQ982718 KYQ51705.1 GANO01000627 JAB59244.1 KQ976606 KYM79325.1 NEVH01013262 PNF29124.1 CAEY01000677 KZ288293 PBC28993.1 GFJQ02001298 JAW05672.1 KK856508 PTY25931.1 CH916366 EDV96290.1 AAGJ04064386 PYGN01000235 PSN50528.1 GGMS01017019 MBY86222.1 CH933809 EDW18598.1 KQ981477 KYN41442.1 KQ414731 KOC62427.1 GFAA01002002 JAU01433.1 QCYY01002113 ROT72779.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000192223 UP000007266 UP000008820 UP000069940 UP000249989 UP000078200 UP000092444 UP000092445 UP000037069 UP000019118 UP000030742 UP000092460 UP000095301 UP000075885 UP000076407 UP000002320 UP000075902 UP000075903 UP000075881 UP000007062 UP000075840 UP000069272 UP000008237 UP000076408 UP000075900 UP000030765 UP000036403 UP000075920 UP000075901 UP000075883 UP000000673 UP000075880 UP000030746 UP000000311 UP000075884 UP000075886 UP000015103 UP000007798 UP000092461 UP000027135 UP000215335 UP000053454 UP000002358 UP000078492 UP000014760 UP000092553 UP000005205 UP000095300 UP000078542 UP000008792 UP000075809 UP000078540 UP000235965 UP000015104 UP000242457 UP000001070 UP000007110 UP000245037 UP000192221 UP000009192 UP000078541 UP000053825 UP000283509
UP000192223 UP000007266 UP000008820 UP000069940 UP000249989 UP000078200 UP000092444 UP000092445 UP000037069 UP000019118 UP000030742 UP000092460 UP000095301 UP000075885 UP000076407 UP000002320 UP000075902 UP000075903 UP000075881 UP000007062 UP000075840 UP000069272 UP000008237 UP000076408 UP000075900 UP000030765 UP000036403 UP000075920 UP000075901 UP000075883 UP000000673 UP000075880 UP000030746 UP000000311 UP000075884 UP000075886 UP000015103 UP000007798 UP000092461 UP000027135 UP000215335 UP000053454 UP000002358 UP000078492 UP000014760 UP000092553 UP000005205 UP000095300 UP000078542 UP000008792 UP000075809 UP000078540 UP000235965 UP000015104 UP000242457 UP000001070 UP000007110 UP000245037 UP000192221 UP000009192 UP000078541 UP000053825 UP000283509
PRIDE
Interpro
SUPFAM
SSF51182
SSF51182
Gene 3D
ProteinModelPortal
H9JMF4
A0A2A4ITF7
A0A2H1VNS3
A0A0N0PEH3
A0A1E1WJG5
A0A194QCL8
+ More
A0A0L7LQX3 A0A1E1W058 A0A212EWC3 A0A1W4WVA0 V5GK94 A0A1Y1JVC2 D6WZW8 Q17FN6 Q16WG8 A0A0K8TL44 A0A0P6IUE8 A0A182GF60 A0A1A9VN32 A0A1B0G3Y8 A0A1A9ZMJ9 A0A0L0C202 A0A1Q3F5R5 N6U292 A0A1B0AQB1 A0A0K8UZC4 A0A1I8MF42 A0A182PIV8 A0A182X277 B0WC79 A0A182U4Z3 A0A182V7B9 A0A182K0P8 A0NBN0 A0A182I6D7 A0A034WKT2 A0A3F2YQA3 E2BKW6 A0A182Y886 A0A1B6L6N8 A0A182R6P9 A0A084WMV9 A0A2M4AZ60 A0A0J7N0L6 A0A182WI33 A0A182SYC0 A0A182MEH2 A0A2M4CP26 A0A1B6GJV6 W5JL03 A0A2M4CNG6 A0A2M4CPI7 A0A1B6JJP4 A0A182JB67 V4BNI4 E2ACF2 A0A182N7F7 A0A182QAW1 A0A0P4VJQ5 R4G7Y4 A0A2P6KAV8 B4MXE0 A0A1B0CQG2 A0A1L8DB51 A0A023F8R6 A0A2S2QMK6 T1DEZ6 A0A1B6CV59 A0A067QV27 A0A232F4D4 A0A0L8HRA0 K7J5B6 A0A195E3R6 R7TVH5 A0A0M4E8X3 A0A158NW35 A0A2R5LIN1 A0A1I8PZM3 A0A195CS53 B4LGZ5 A0A151WVK3 U5EWC7 A0A151I0T0 A0A2J7QKL0 T1KXA8 A0A2A3EB42 A0A1Z5LE34 A0A2R7X2P3 B4IWR4 W4YHW1 A0A2P8Z212 A0A1W4VNG9 A0A2S2RA44 B4KWX7 A0A151JYD8 A0A0L7QUU3 A0A1E1XQ38 A0A3R7QAD7
A0A0L7LQX3 A0A1E1W058 A0A212EWC3 A0A1W4WVA0 V5GK94 A0A1Y1JVC2 D6WZW8 Q17FN6 Q16WG8 A0A0K8TL44 A0A0P6IUE8 A0A182GF60 A0A1A9VN32 A0A1B0G3Y8 A0A1A9ZMJ9 A0A0L0C202 A0A1Q3F5R5 N6U292 A0A1B0AQB1 A0A0K8UZC4 A0A1I8MF42 A0A182PIV8 A0A182X277 B0WC79 A0A182U4Z3 A0A182V7B9 A0A182K0P8 A0NBN0 A0A182I6D7 A0A034WKT2 A0A3F2YQA3 E2BKW6 A0A182Y886 A0A1B6L6N8 A0A182R6P9 A0A084WMV9 A0A2M4AZ60 A0A0J7N0L6 A0A182WI33 A0A182SYC0 A0A182MEH2 A0A2M4CP26 A0A1B6GJV6 W5JL03 A0A2M4CNG6 A0A2M4CPI7 A0A1B6JJP4 A0A182JB67 V4BNI4 E2ACF2 A0A182N7F7 A0A182QAW1 A0A0P4VJQ5 R4G7Y4 A0A2P6KAV8 B4MXE0 A0A1B0CQG2 A0A1L8DB51 A0A023F8R6 A0A2S2QMK6 T1DEZ6 A0A1B6CV59 A0A067QV27 A0A232F4D4 A0A0L8HRA0 K7J5B6 A0A195E3R6 R7TVH5 A0A0M4E8X3 A0A158NW35 A0A2R5LIN1 A0A1I8PZM3 A0A195CS53 B4LGZ5 A0A151WVK3 U5EWC7 A0A151I0T0 A0A2J7QKL0 T1KXA8 A0A2A3EB42 A0A1Z5LE34 A0A2R7X2P3 B4IWR4 W4YHW1 A0A2P8Z212 A0A1W4VNG9 A0A2S2RA44 B4KWX7 A0A151JYD8 A0A0L7QUU3 A0A1E1XQ38 A0A3R7QAD7
Ontologies
KEGG
PATHWAY
GO
PANTHER
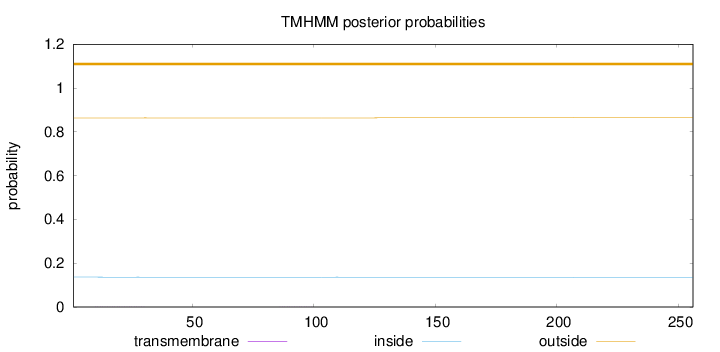
Topology
Length:
256
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02278
Exp number, first 60 AAs:
0.01801
Total prob of N-in:
0.13679
outside
1 - 256
Population Genetic Test Statistics
Pi
215.045639
Theta
232.991088
Tajima's D
-0.184586
CLR
386.489804
CSRT
0.31868406579671
Interpretation
Uncertain
