Pre Gene Modal
BGIBMGA010847
Annotation
N-phosphoacetylglucosamine_mutase_[Cnaphalocrocis_medinalis]
Full name
Phosphoacetylglucosamine mutase
Alternative Name
Acetylglucosamine phosphomutase
N-acetylglucosamine-phosphate mutase
N-acetylglucosamine-phosphate mutase
Location in the cell
Cytoplasmic Reliability : 2.927
Sequence
CDS
ATGCCGCCGAGTAGCTTACGAGTCGTGTACGCGTTCGCCAGAGAAATGCATCCGAAGACAACTGACATTTTTATTGAATACGGCACTGCCGGATTTCGTACAAAGGCCAATTTATTGGAGCATGTCGTGTATCGTATGGGTCTCTTGGCTGTCATTCGTTCCAGAGTTAAAAATGGACGCACCATCGGCATCATGATAACAGCATCGCACAATCTCGAGCCCGACAACGGTGTGAAGCTCATCGATCCAGATGGCGAGATGCTTGAGCAGAGCTGGGAGGCGATAGCCACTAAACTAGCTAATGTCAGTGACAATGACTTGGAAGCGACAACTGCTGAGATTATAAAAGAAGTGAATGCTAATATGACACTCAAAACGAGTATATTCATTGGAATGGACACAAGATACACAAGTCCCCGTCTAGCACAGGCGGCTGCTAATGGAGTCATGGCGCTGAAGGGAACGCCCAAGGAGTTCGGAATTGTTACCACTCCAATGCTGCATTTCTTCGTAAAATGTCGTAACGATAATACATATGGAACTCCAACCGAGGAAGGATATTATGAGAAAATCGTTGGTGCTTTCAAGAATATACGTCAAAAACTACCCGTTTACGGTAATTATAGTACAACGTTGTACGTAGACGGCGCTAACGGCGTCGGTGGAAAGAAACTTAATATCATCAAGAAGACCCTCGATGGTGAATTGGATCTGAAGCTGTTCAATTTAGGCGGGAACGGAGGAAAGTTGAATTTGAATTGCGGTGCTGATTTCGTGAAAGTATCACAGAAGCCGCCCGTCGGCGTAGAGCACGTGCCGATGCAGCGAGTGGCCTCACTGGACGGGGACTCCGATCGCATCGTCTATTACTACGTCGACGATAAAGATAAAATGCACTTGTTGGACGGCGATCGTATAGCCACATTACTGGCCAGCTACATCACCGAATTGCTGACGGCCAGCGAAGCGAAACATTTGAAACTAGGCCTCGTGCAAACGGCCTACGCTAACGGTGCATCCACTGCTTATATAACGCAGAAGCTGAAAGTCCCGGTGAGCTGCGTGAAGACAGGAGTGAAGCATCTCCATCACGCAGCGCTTTCGTATGACATCGGCGTCTACTTTGAAGCGAACGGGCACGGCACAGTCATATACAGTCACGACGCTAAGAAGACGATCAGCAAAATTGCCGAAGAGGGGGAGTCGGAACAAAGGAAGGCGGCGCAGTTGCTTCTGGACTTCATCGACATGACCAACGAGACGGTCGGAGACGCCATATCGGACCTGTTCCTCGTCGAGACGGTGCTCTGCGCTCGAGGACTGGACGCCGAGCAGTGGCTCTCCACGTACGACGACCTGCCCTGCAGACAAGTAAAAGTAACAGTACAGGATCGTAACGTGATATCTACGGCGGACGCGGAGCGCGTGTGCACGTCGCCCGAGGGACTGCAGACGCGTATCGACGAGCTCGTCGCTGCGTACGCGGACGGACGCGCCTTCGTGCGTCCCTCCGGCACCGAGGACGTGGTGCGAGTGTACGCGGAAGCGGATACGCAGCAGTCAGCAGATAAACTCGCCGCAGAAGTATCACAGGCCGTTTACGATTTGGCCGGCGGCGTTGGAAATCGGCCAGAGTTGCCAGCTTAA
Protein
MPPSSLRVVYAFAREMHPKTTDIFIEYGTAGFRTKANLLEHVVYRMGLLAVIRSRVKNGRTIGIMITASHNLEPDNGVKLIDPDGEMLEQSWEAIATKLANVSDNDLEATTAEIIKEVNANMTLKTSIFIGMDTRYTSPRLAQAAANGVMALKGTPKEFGIVTTPMLHFFVKCRNDNTYGTPTEEGYYEKIVGAFKNIRQKLPVYGNYSTTLYVDGANGVGGKKLNIIKKTLDGELDLKLFNLGGNGGKLNLNCGADFVKVSQKPPVGVEHVPMQRVASLDGDSDRIVYYYVDDKDKMHLLDGDRIATLLASYITELLTASEAKHLKLGLVQTAYANGASTAYITQKLKVPVSCVKTGVKHLHHAALSYDIGVYFEANGHGTVIYSHDAKKTISKIAEEGESEQRKAAQLLLDFIDMTNETVGDAISDLFLVETVLCARGLDAEQWLSTYDDLPCRQVKVTVQDRNVISTADAERVCTSPEGLQTRIDELVAAYADGRAFVRPSGTEDVVRVYAEADTQQSADKLAAEVSQAVYDLAGGVGNRPELPA
Summary
Description
Catalyzes the conversion of GlcNAc-6-P into GlcNAc-1-P during the synthesis of uridine diphosphate/UDP-GlcNAc, a sugar nucleotide critical to multiple glycosylation pathways including protein N- and O-glycosylation.
Catalytic Activity
N-acetyl-alpha-D-glucosamine 1-phosphate = N-acetyl-D-glucosamine 6-phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the phosphohexose mutase family.
Uniprot
A0A1C9CW41
A0A3S2M0H4
A0A0G2RLT8
A0A194QC94
A0A2A4JC27
A0A212EW76
+ More
A0A0N0PE63 A0A2H1VM40 B4PGS6 V5GWV2 B3NHC2 A0A1Y1M850 Q9VTZ4 A0A1W4UGY7 D6X3A7 B4QRP0 A0A1B0CKT3 A0A0T6B8G5 A0A1L8E2L9 A0A336LSL1 Q2M0L7 A0A336LW80 B3MB25 A0A3B0KMN4 A0A1W4WEH6 A0A1W4W2M4 A0A336M7Q0 B4L0Q6 B4N332 B4LGP2 Q16PT5 A0A182GUX6 B4IX69 A0A3B0JSG8 B4NMG7 Q58I85 A0A1I8NN57 A0A0K8VIJ8 B0WVX0 W8BLH0 A0A1Q3F954 A0A1Q3F971 A0A0M5J3X5 B0X482 A0A034VJK5 A0A1J1IP77 A0A0A1XQ05 A0A0L0CL80 A0A182R3J0 A0A182JC50 A0A182PP35 A0A182WJR6 A0A182MN30 A0A084WES1 A0A182YML6 A0A182KQN4 A0A2C9GRT4 A0A182TJB0 Q7QJ25 A0A182VJ82 T1PG23 A0A1I8NAA5 A0A182N917 A0A2Z4BZJ4 D3TNI4 A0A1A9UTQ9 A0A182QQ20 A0A1B0GA53 A0A182FGD0 W5J6K1 A0A182XLJ6 K7J1K6 A0A1B6FN26 B4GRG5 E9GZ07 A0A067RFT3 A0A0P5VCP7 A0A0P6AG10 A0A0P5SY06 A0A0P5WWJ7 A0A0P5TZI9 A0A1B0AR70 A0A0P6IC12 A0A0P5VVA3 A0A0P5UWV5 A0A0P5UWP2 A0A0P4ZCJ5 A0A1B6KB84 A0A0P4YNT6 A0A0P5NPK9 A0A3R7PN50 R7UDD6 A0A0N7ZZG9 A0A1S3IPZ1 A0A3G2KJL5 A0A3P8WV03 G3WU51 K1QVE8 A0A2J7QDT1 A0A3B4ZQD2
A0A0N0PE63 A0A2H1VM40 B4PGS6 V5GWV2 B3NHC2 A0A1Y1M850 Q9VTZ4 A0A1W4UGY7 D6X3A7 B4QRP0 A0A1B0CKT3 A0A0T6B8G5 A0A1L8E2L9 A0A336LSL1 Q2M0L7 A0A336LW80 B3MB25 A0A3B0KMN4 A0A1W4WEH6 A0A1W4W2M4 A0A336M7Q0 B4L0Q6 B4N332 B4LGP2 Q16PT5 A0A182GUX6 B4IX69 A0A3B0JSG8 B4NMG7 Q58I85 A0A1I8NN57 A0A0K8VIJ8 B0WVX0 W8BLH0 A0A1Q3F954 A0A1Q3F971 A0A0M5J3X5 B0X482 A0A034VJK5 A0A1J1IP77 A0A0A1XQ05 A0A0L0CL80 A0A182R3J0 A0A182JC50 A0A182PP35 A0A182WJR6 A0A182MN30 A0A084WES1 A0A182YML6 A0A182KQN4 A0A2C9GRT4 A0A182TJB0 Q7QJ25 A0A182VJ82 T1PG23 A0A1I8NAA5 A0A182N917 A0A2Z4BZJ4 D3TNI4 A0A1A9UTQ9 A0A182QQ20 A0A1B0GA53 A0A182FGD0 W5J6K1 A0A182XLJ6 K7J1K6 A0A1B6FN26 B4GRG5 E9GZ07 A0A067RFT3 A0A0P5VCP7 A0A0P6AG10 A0A0P5SY06 A0A0P5WWJ7 A0A0P5TZI9 A0A1B0AR70 A0A0P6IC12 A0A0P5VVA3 A0A0P5UWV5 A0A0P5UWP2 A0A0P4ZCJ5 A0A1B6KB84 A0A0P4YNT6 A0A0P5NPK9 A0A3R7PN50 R7UDD6 A0A0N7ZZG9 A0A1S3IPZ1 A0A3G2KJL5 A0A3P8WV03 G3WU51 K1QVE8 A0A2J7QDT1 A0A3B4ZQD2
EC Number
5.4.2.3
Pubmed
26354079
22118469
17994087
17550304
28004739
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 22936249 15632085 17510324 26483478 24495485 25348373 25830018 26108605 24438588 25244985 20966253 12364791 14747013 17210077 25315136 20353571 20920257 23761445 20075255 21292972 24845553 23254933 26383154 30326269 24487278 21709235 22992520
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 22936249 15632085 17510324 26483478 24495485 25348373 25830018 26108605 24438588 25244985 20966253 12364791 14747013 17210077 25315136 20353571 20920257 23761445 20075255 21292972 24845553 23254933 26383154 30326269 24487278 21709235 22992520
EMBL
KX130001
AON76440.1
RSAL01000087
RVE48266.1
KP000845
AJG44540.1
+ More
KQ459193 KPJ03163.1 NWSH01002183 PCG68953.1 AGBW02012063 OWR45745.1 KQ459941 KPJ19152.1 ODYU01003314 SOQ41905.1 CM000159 EDW94315.1 GALX01003713 JAB64753.1 CH954178 EDV51579.1 GEZM01037864 JAV81873.1 AE014296 AY069309 AAF49901.1 AAL39454.1 KQ971372 EFA09778.2 CM000363 CM002912 EDX10254.1 KMY99259.1 AJWK01016608 LJIG01009161 KRT83634.1 GFDF01001339 JAV12745.1 UFQS01000155 UFQT01000155 SSX00594.1 SSX20974.1 CH379069 EAL30913.1 UFQT01000150 SSX20913.1 CH902618 EDV40291.1 OUUW01000009 SPP85068.1 UFQT01000659 SSX26235.1 CH933809 EDW18133.1 CH964062 EDW78771.1 CH940647 EDW70507.1 CH477773 EAT36375.1 JXUM01018692 JXUM01018693 KQ560519 KXJ82014.1 CH916366 EDV97401.1 SPP85067.1 CH964282 EDW85556.1 AY944754 AAX47077.1 GDHF01019812 GDHF01013949 JAI32502.1 JAI38365.1 DS232134 EDS35802.1 GAMC01006898 JAB99657.1 GFDL01010940 JAV24105.1 GFDL01010914 JAV24131.1 CP012525 ALC44344.1 DS232331 EDS40159.1 GAKP01017259 GAKP01017258 JAC41693.1 CVRI01000054 CRL00297.1 GBXI01001347 GBXI01001164 JAD12945.1 JAD13128.1 JRES01000240 KNC33055.1 AXCM01002191 ATLV01023246 KE525341 KFB48715.1 APCN01000178 AAAB01008807 EAA04228.3 KA647767 AFP62396.1 MH119583 AWU68201.1 EZ422986 ADD19262.1 AXCN02000540 CCAG010015850 ADMH02002070 ETN59626.1 GECZ01018208 JAS51561.1 CH479188 EDW40350.1 GL732576 EFX75314.1 KK852714 KDR17892.1 GDIP01102141 JAM01574.1 GDIP01043245 LRGB01000687 JAM60470.1 KZS16738.1 GDIP01138493 JAL65221.1 GDIP01080792 JAM22923.1 GDIP01137188 JAL66526.1 JXJN01002268 GDIQ01023112 JAN71625.1 GDIP01095800 JAM07915.1 GDIP01107588 JAL96126.1 GDIP01107589 JAL96125.1 GDIP01217477 GDIP01215078 JAJ08324.1 GEBQ01031264 JAT08713.1 GDIP01224575 JAI98826.1 GDIQ01139576 JAL12150.1 QCYY01002239 ROT71834.1 AMQN01008178 KB302363 ELU04395.1 GDIP01197385 JAJ26017.1 MH320769 AYN59160.1 AEFK01168771 JH818113 EKC40852.1 NEVH01015814 PNF26738.1
KQ459193 KPJ03163.1 NWSH01002183 PCG68953.1 AGBW02012063 OWR45745.1 KQ459941 KPJ19152.1 ODYU01003314 SOQ41905.1 CM000159 EDW94315.1 GALX01003713 JAB64753.1 CH954178 EDV51579.1 GEZM01037864 JAV81873.1 AE014296 AY069309 AAF49901.1 AAL39454.1 KQ971372 EFA09778.2 CM000363 CM002912 EDX10254.1 KMY99259.1 AJWK01016608 LJIG01009161 KRT83634.1 GFDF01001339 JAV12745.1 UFQS01000155 UFQT01000155 SSX00594.1 SSX20974.1 CH379069 EAL30913.1 UFQT01000150 SSX20913.1 CH902618 EDV40291.1 OUUW01000009 SPP85068.1 UFQT01000659 SSX26235.1 CH933809 EDW18133.1 CH964062 EDW78771.1 CH940647 EDW70507.1 CH477773 EAT36375.1 JXUM01018692 JXUM01018693 KQ560519 KXJ82014.1 CH916366 EDV97401.1 SPP85067.1 CH964282 EDW85556.1 AY944754 AAX47077.1 GDHF01019812 GDHF01013949 JAI32502.1 JAI38365.1 DS232134 EDS35802.1 GAMC01006898 JAB99657.1 GFDL01010940 JAV24105.1 GFDL01010914 JAV24131.1 CP012525 ALC44344.1 DS232331 EDS40159.1 GAKP01017259 GAKP01017258 JAC41693.1 CVRI01000054 CRL00297.1 GBXI01001347 GBXI01001164 JAD12945.1 JAD13128.1 JRES01000240 KNC33055.1 AXCM01002191 ATLV01023246 KE525341 KFB48715.1 APCN01000178 AAAB01008807 EAA04228.3 KA647767 AFP62396.1 MH119583 AWU68201.1 EZ422986 ADD19262.1 AXCN02000540 CCAG010015850 ADMH02002070 ETN59626.1 GECZ01018208 JAS51561.1 CH479188 EDW40350.1 GL732576 EFX75314.1 KK852714 KDR17892.1 GDIP01102141 JAM01574.1 GDIP01043245 LRGB01000687 JAM60470.1 KZS16738.1 GDIP01138493 JAL65221.1 GDIP01080792 JAM22923.1 GDIP01137188 JAL66526.1 JXJN01002268 GDIQ01023112 JAN71625.1 GDIP01095800 JAM07915.1 GDIP01107588 JAL96126.1 GDIP01107589 JAL96125.1 GDIP01217477 GDIP01215078 JAJ08324.1 GEBQ01031264 JAT08713.1 GDIP01224575 JAI98826.1 GDIQ01139576 JAL12150.1 QCYY01002239 ROT71834.1 AMQN01008178 KB302363 ELU04395.1 GDIP01197385 JAJ26017.1 MH320769 AYN59160.1 AEFK01168771 JH818113 EKC40852.1 NEVH01015814 PNF26738.1
Proteomes
UP000283053
UP000053268
UP000218220
UP000007151
UP000053240
UP000002282
+ More
UP000008711 UP000000803 UP000192221 UP000007266 UP000000304 UP000092461 UP000001819 UP000007801 UP000268350 UP000192223 UP000009192 UP000007798 UP000008792 UP000008820 UP000069940 UP000249989 UP000001070 UP000095300 UP000002320 UP000092553 UP000183832 UP000037069 UP000075900 UP000075880 UP000075885 UP000075920 UP000075883 UP000030765 UP000076408 UP000075882 UP000075840 UP000075902 UP000007062 UP000075903 UP000095301 UP000075884 UP000078200 UP000075886 UP000092444 UP000069272 UP000000673 UP000076407 UP000002358 UP000008744 UP000000305 UP000027135 UP000076858 UP000092460 UP000283509 UP000014760 UP000085678 UP000265120 UP000007648 UP000005408 UP000235965 UP000261400
UP000008711 UP000000803 UP000192221 UP000007266 UP000000304 UP000092461 UP000001819 UP000007801 UP000268350 UP000192223 UP000009192 UP000007798 UP000008792 UP000008820 UP000069940 UP000249989 UP000001070 UP000095300 UP000002320 UP000092553 UP000183832 UP000037069 UP000075900 UP000075880 UP000075885 UP000075920 UP000075883 UP000030765 UP000076408 UP000075882 UP000075840 UP000075902 UP000007062 UP000075903 UP000095301 UP000075884 UP000078200 UP000075886 UP000092444 UP000069272 UP000000673 UP000076407 UP000002358 UP000008744 UP000000305 UP000027135 UP000076858 UP000092460 UP000283509 UP000014760 UP000085678 UP000265120 UP000007648 UP000005408 UP000235965 UP000261400
PRIDE
Pfam
Interpro
IPR005843
A-D-PHexomutase_C
+ More
IPR016055 A-D-PHexomutase_a/b/a-I/II/III
IPR036900 A-D-PHexomutase_C_sf
IPR005844 A-D-PHexomutase_a/b/a-I
IPR016657 PAGM
IPR016066 A-D-PHexomutase_CS
IPR005846 A-D-PHexomutase_a/b/a-III
IPR027974 DUF4470
IPR028235 DNAAF3_C
IPR015915 Kelch-typ_b-propeller
IPR018195 Transferrin_Fe_BS
IPR001156 Transferrin-like_dom
IPR016055 A-D-PHexomutase_a/b/a-I/II/III
IPR036900 A-D-PHexomutase_C_sf
IPR005844 A-D-PHexomutase_a/b/a-I
IPR016657 PAGM
IPR016066 A-D-PHexomutase_CS
IPR005846 A-D-PHexomutase_a/b/a-III
IPR027974 DUF4470
IPR028235 DNAAF3_C
IPR015915 Kelch-typ_b-propeller
IPR018195 Transferrin_Fe_BS
IPR001156 Transferrin-like_dom
Gene 3D
CDD
ProteinModelPortal
A0A1C9CW41
A0A3S2M0H4
A0A0G2RLT8
A0A194QC94
A0A2A4JC27
A0A212EW76
+ More
A0A0N0PE63 A0A2H1VM40 B4PGS6 V5GWV2 B3NHC2 A0A1Y1M850 Q9VTZ4 A0A1W4UGY7 D6X3A7 B4QRP0 A0A1B0CKT3 A0A0T6B8G5 A0A1L8E2L9 A0A336LSL1 Q2M0L7 A0A336LW80 B3MB25 A0A3B0KMN4 A0A1W4WEH6 A0A1W4W2M4 A0A336M7Q0 B4L0Q6 B4N332 B4LGP2 Q16PT5 A0A182GUX6 B4IX69 A0A3B0JSG8 B4NMG7 Q58I85 A0A1I8NN57 A0A0K8VIJ8 B0WVX0 W8BLH0 A0A1Q3F954 A0A1Q3F971 A0A0M5J3X5 B0X482 A0A034VJK5 A0A1J1IP77 A0A0A1XQ05 A0A0L0CL80 A0A182R3J0 A0A182JC50 A0A182PP35 A0A182WJR6 A0A182MN30 A0A084WES1 A0A182YML6 A0A182KQN4 A0A2C9GRT4 A0A182TJB0 Q7QJ25 A0A182VJ82 T1PG23 A0A1I8NAA5 A0A182N917 A0A2Z4BZJ4 D3TNI4 A0A1A9UTQ9 A0A182QQ20 A0A1B0GA53 A0A182FGD0 W5J6K1 A0A182XLJ6 K7J1K6 A0A1B6FN26 B4GRG5 E9GZ07 A0A067RFT3 A0A0P5VCP7 A0A0P6AG10 A0A0P5SY06 A0A0P5WWJ7 A0A0P5TZI9 A0A1B0AR70 A0A0P6IC12 A0A0P5VVA3 A0A0P5UWV5 A0A0P5UWP2 A0A0P4ZCJ5 A0A1B6KB84 A0A0P4YNT6 A0A0P5NPK9 A0A3R7PN50 R7UDD6 A0A0N7ZZG9 A0A1S3IPZ1 A0A3G2KJL5 A0A3P8WV03 G3WU51 K1QVE8 A0A2J7QDT1 A0A3B4ZQD2
A0A0N0PE63 A0A2H1VM40 B4PGS6 V5GWV2 B3NHC2 A0A1Y1M850 Q9VTZ4 A0A1W4UGY7 D6X3A7 B4QRP0 A0A1B0CKT3 A0A0T6B8G5 A0A1L8E2L9 A0A336LSL1 Q2M0L7 A0A336LW80 B3MB25 A0A3B0KMN4 A0A1W4WEH6 A0A1W4W2M4 A0A336M7Q0 B4L0Q6 B4N332 B4LGP2 Q16PT5 A0A182GUX6 B4IX69 A0A3B0JSG8 B4NMG7 Q58I85 A0A1I8NN57 A0A0K8VIJ8 B0WVX0 W8BLH0 A0A1Q3F954 A0A1Q3F971 A0A0M5J3X5 B0X482 A0A034VJK5 A0A1J1IP77 A0A0A1XQ05 A0A0L0CL80 A0A182R3J0 A0A182JC50 A0A182PP35 A0A182WJR6 A0A182MN30 A0A084WES1 A0A182YML6 A0A182KQN4 A0A2C9GRT4 A0A182TJB0 Q7QJ25 A0A182VJ82 T1PG23 A0A1I8NAA5 A0A182N917 A0A2Z4BZJ4 D3TNI4 A0A1A9UTQ9 A0A182QQ20 A0A1B0GA53 A0A182FGD0 W5J6K1 A0A182XLJ6 K7J1K6 A0A1B6FN26 B4GRG5 E9GZ07 A0A067RFT3 A0A0P5VCP7 A0A0P6AG10 A0A0P5SY06 A0A0P5WWJ7 A0A0P5TZI9 A0A1B0AR70 A0A0P6IC12 A0A0P5VVA3 A0A0P5UWV5 A0A0P5UWP2 A0A0P4ZCJ5 A0A1B6KB84 A0A0P4YNT6 A0A0P5NPK9 A0A3R7PN50 R7UDD6 A0A0N7ZZG9 A0A1S3IPZ1 A0A3G2KJL5 A0A3P8WV03 G3WU51 K1QVE8 A0A2J7QDT1 A0A3B4ZQD2
PDB
5OAW
E-value=6.21032e-128,
Score=1173
Ontologies
PATHWAY
GO
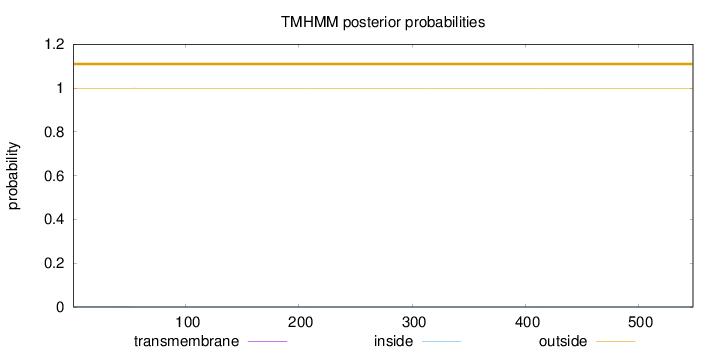
Topology
Length:
548
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00523999999999998
Exp number, first 60 AAs:
0.00283
Total prob of N-in:
0.00064
outside
1 - 548
Population Genetic Test Statistics
Pi
179.617485
Theta
154.803411
Tajima's D
0.88129
CLR
1.93603
CSRT
0.623868806559672
Interpretation
Uncertain
