Pre Gene Modal
BGIBMGA010711
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_polybromo-1_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.242
Sequence
CDS
ATGAGCAAGAGACGCAGAGCGTCTTCTGGGGCAAGCCGAGCTGATGGTGACGATAGTGATGAACTAAGTAATCCTGGACCACCACCATCGAACAGGAAGAGGAAAAAGTTGGATCCTAGTGAAATTTGCCAGCAACTTTATGATACAATAAGATCTCATAAGAAAGAAGACGGGACTCTGTTATGTGACTCATTTATAAGGGCTCCAAAGAGACGACAGGAACCTCAGTACTACGAAGTTGTATCACAACCAATTGATTTACTTAGAGTACAACAAAAACTTAAAACAGACACATATGAAGACATTGAAGAATTAACAGCAGATATAGAGCTACTGGTGAACAATGCCAAAGCATTCTACAAACCTGACACTGTTGAGTATAAAGATGCAGTTGACCTTTGGAAGTTATTCTTGCATACTAAACAAACACTGGAACATGATGAAGACATGAAAACTCCTAAATTATCACGAAATGGTTCATCAAGTAGGAGATCAGATGTGGCTGAAGATTTGTCTGAAACATCCACTAACAATGAAGAAGACAATGTTTTTGAAGAGCTATTCAGTGCGGTTATGACTGCGAACATTGATGGACGCCCACTTTACCAAGCATTTCAATTTATACCTTCGAAGCGGCGTTACCCTGAGTACTTTAATGTGATTGATTCGCCTATTGACCTAAAGACGATAGCACAGAAGATTCAAGGCGGAGAATACACTACTATTGGGGAATTAGAAAAAGACTTCCTACTCATGGTCAGAAACGCCTGTCAGTTCAACGAGCCTGGAAGTCAGATTTATAAAGATGCCAAAACTTTAAAGAAAATAATACAAGTACGCAGACAAGAAATCGAGCAGCACGGTCGCAGTGGACCCGCCAAAACATCGGAACGCATACGGAGCAAACGTACATCTAGAGTCGGACCGGTGCCCACTAGTAGGGCTCTGGCCATCATGGAACCGCCCGGATCCGATACTGAAGCCTATGTCAAGCATTCTGAAGATAGCGCTGAAAGTGATGAAGATAAAGTGGATAATGAAGATTCTCCACAGTGGAAGTTGTTGGAGACTGTCAAGAACCATCTTGGTCCTAGTGGGTTACCAATGGCCGAACCATTTTGGAAATTACCATCTCGTCGCGCCTACCCTAATTATTACAAGGAGATAAAGAACCCTGTGTCTTTAAACCAAATCAAAAACAAAATACGACGTGGCAGCTACGGTACTCTGTCCGAGGTGGCGGGTGATATGAATATTATGTTTGAGAACGCCAAACAGTACAACGCCCCCACTTCCAGACTATATAAGGACTCTGTAAAATTGCAACGCTTAATGCAACAACGTGTCCAAGAACTCCTGGATATAGTTCAGTCATCATCGAGTGATGATGAAAGCCTGTCTTCAGTAAAGAATCAAACTCAAGTGCAGACTCCACGGCCAAGAGGTCGTCCGCGCATCAACCCGCTGCCCGCGCCCGCCCCGCCGCCCGCCGCCGCGCCGCTCGTGCTGAAGCCGAACATGCCGCTGAAGAAGAAGCTGCACTACGTGGCCAAGCAGCTGGTCGAGTTCACGTGCTCCGACGGCCGCCAGCCGATGCTGCTGTTCATGGAAAAGCCCAGCAAGAAACTGTATCCGGAATATTACAACGTCATCGATCGGCCCATCGATATGCTGACGATTGAAGCTAACATTAAGAACGATCGTTATAATTCACTAGAAGAGATGATAGCGGACTTCCGTTTGATGTTCTCAAACTGCCGCCATTTCAACGAAGAGGGCTCTATGGTATACGAGGACGCGAACCTTCTAGAACGGGTCATGAACGAAAAGATAAAGGAATTGAACAGCAATTACGAGAAAAAGACGCCCATGAAGAGTCTGAAGGCTCAGCCAAGGTCGAGGCAACTGTCGCCGTTCGAACAGAAGCTGAGAACGTTGTATGACGCTATCCGGGACTATAGGGATCCAAAAGCCAACCGCCAGCTGGCCCTGATCTTCATGAAGCTACCCAGTAAAACTGAGTACCCTGATTATTATGAGCTCATCAAGAACCCGATAGACATGGAGAAGATTGCTCACAAATTGAAGAGCAACAGTTACAGTTCAGTTAACGAGCTGGCTTCGGATTTCATACTGATGTTCGATAACGCCTGCAAGTACAATGAGCCCGACTCGCAGATCTACAAGGACGCTTTGATACTGCACAGAGTCTGTCTGCAGACCAAGCAGATGTTAAGTCAAGACGACGATGCAGTGCCCGACGTTCCAGCCGCTGTCCAAGAACTATTATTGAATCTCTTTACCACAGTATACAATCATCAAGATGAAGAGGGGAGGTGTTACTCAGACAGCATGGCCGAGTTACCGGAACACGACGAAACAAACAATGGCGAGAAGATGCGAGCTATATCGCTGGATTTGGTCAAGAGAAGACTTGACAAAGGGCTGTACAAGAGACTGGATCATTTCCAGCAAGATATGTTTGCTGTGTTTGAAAGAGCACGTCGCCTCTCCCGTACAGATAGTCAAATTTTCGAAGACTCAGTCGAGCTGCAGTGCTACTACATAGAGCAGAGGGACGCGCTGTGCCGCGGCACGCTGCAGTCGCCGGCGCTCACCTTCACGCGCGACACCGTCTCCACCAGCGTCGAGCTCATCAAGCAGTGCAAGCTGCTGCAAGAGAACGACGATGAGGATGAAACTAGATCGAGTACGGATGACTCCATGCCGTCGGGTGATACCAATCTTCAATCTCAGTACAGGAAAGGTGATTTCCTGTACATCCAACCTGAGAAGGGCAATAAAGACTTCAATATTGTTCAAGTAGAAAGACTTTGGACTAACAGCGATGGTATAACTATGCTGTATTGCAACATGTACTTTAGACCTCAAGAGACATTCCACATCAGAACCCGAAAGTTCCTCCAACAAGAAGTGTTCAAAACGGATGTGTACAAACCTGTGCCCCTGGACCGCGTCGTGGGACCTTGCTACGTTATGAACGTGAAGGAATATTTCAAGTTTAGACCTGAAGGCTACGCTGACAAAGACGTCTACGTCTGCGAGAGTCGGTACAGCACGAAGCACAGGTGGTTCAAAAAGATCAGAGCCTGGGAGGGCTCCGAGAAAGAAGTGTCGATTGTTCCCAGAGAGGTGGCGCTAGAGCCGCAGCGGACCGTCTCCGTGTTCCGCGAGCGCGTCGAGAAGCACAAAGACGAGCTGGCCGAGCTAGAAGTACTCGAGAACGTGCAAGAAAAGGAGAGGCCGGACGTGGTGATGTACAACCCGCTGGGCACGGACGACGAGAACACGTACTACGAGCAGTACAACACGGTGTGCTCGGGCGTCATCAAGACCGGCGACTACGTGTACGTGGTCACGGACGGCGGGAAGCAGATGATAGCGCAGGTCGACACCATCTGGGAGACCGGCGATAACAAGTGCTATTTCCGTGGACCGTTCTTGATCTTCCCGGCGGAGGTGTTGAATATCATCAATAAGCCTTTCTACAAACAAGAGGTACTTCTGACAACAATCCACGATACTAGCCCTCTTGTGGGCATCGTTGGGAAGTGTTCGGTTTTGGACTACGACGATTATCTCAAATGTCGGCCCACGGAGATATCGGAAGGCGACGTGTACGTGTGCGAGAGCATCTACGACGAGTCGAACCGCGTCGCGCGCAAGCTCAAGTCCGGCCTGCGCAAGTTCGAGCACACCAAGGACGTCACCGTAGACGAGATATACTTCTTCCCAAAACCGTTGGGTCCACCGGCCCTGGCGTCCACGCACGACGTGCAGTCGGCGTCCACCGCCTTCGCCCAGAAACCGCAACATCAAACCATGCACATGGATCCTACGGACTGCAAGCCACAGTTCACGAACCTAATCAACACGACGATCGGAAGTCAGGACGTCGAAATGATTCTAGAGAATTCGTTGGACGACTCGTCTTTGGCCTCGCCGGCGACACCTCTGTCAACAGGTGGCAATAGCAACCCTTACAACCCGTCCCTGAGCGTAACGCCCAGTCAGGAACGGATCGCAAGCCAGACCGCGACCCCGGTCACCGGGAAGAAGAAGAAGGAACAGAAGCAAAAGATCGTCACCGGCTACATACTGTACTCCAGCGAGGTGCGGAAGGCGGTCATAGCCAACAACCCGGAGGCTACATTCGGCGACATATCGCGCATCGTGGGCAACGAGTGGCGCTCGCTGCCCGCCGCCACCAAGCAGAGCTGGGAGGAGCGAGCCGCGCGCTGCAACGAGGAGACCGCCGCCAGGCTCGCCGACGAGATGCGCGAGTTGGCGCAACACACAACAATGGAGATGACGTACGAATGCGCCTGGGACACGTGCGACTGTCAGTTCGAGGACCTGACCGACTGCATGGAGCACTGCATCGGCGACGGAGGAAACACGGGGCACATCCAGCAACACTACCGCGGCTCGTTCAGCGAGTACCCCTGTCTGTGGCGGAACTGCGCGCGCGTGCGCAAGGGGCAGGCGCCCTTCCCCAACCTGCCGCGCCTGCTGCGACACGTGCGCGACCTGCACGTCAACAAGGGCAACGGACGACTCATGGCCGTGCATGAACGCTCCAGGAATTTCATGCTGTCATCGAAGAAGCCGCGGCCGGCGGTGCGGAGCGGCGTCATGTCCCCCGGAGGTACAAATAGCAGTAGTAATAGCCAAGCGTCTCTATCAGGGATGTCCCCTATGGCGAGGAACACCCCGTCCCCCGGCGCGGGGGAGGGCACGTCCACGACGATCCCCACGGCGCCGCCCGTCCGCGCCGGCCTCGACCCCCTCTTCGTTACGGCGCCGCCCAGAGCGCAGCGCGTCACGCACTCCGAAGCCTACATCAGGTACATCGAAGGCTTGCATTCAGAGCAGAAATATATAACACCATGGGAAAAATCTCTTACTCCGATGCCGGCGAACCCCGACCCGGCTCAGTTCAATATGCAGAAGATACCCGGACATTGGATCACGGACGAAGCCATCAACGGATATTTGACACATGACAAAAATCTCGCCGAAGCGGACATACAGAAGATGGACCAAAATCAAAAGGTTCTGAAAGGATTGTGCTCGTTGAGAGATTTCATGATGAAAGACGCCTTGTGTCTCTACAAGAATCTTCAGATAAATGGTTTTTAA
Protein
MSKRRRASSGASRADGDDSDELSNPGPPPSNRKRKKLDPSEICQQLYDTIRSHKKEDGTLLCDSFIRAPKRRQEPQYYEVVSQPIDLLRVQQKLKTDTYEDIEELTADIELLVNNAKAFYKPDTVEYKDAVDLWKLFLHTKQTLEHDEDMKTPKLSRNGSSSRRSDVAEDLSETSTNNEEDNVFEELFSAVMTANIDGRPLYQAFQFIPSKRRYPEYFNVIDSPIDLKTIAQKIQGGEYTTIGELEKDFLLMVRNACQFNEPGSQIYKDAKTLKKIIQVRRQEIEQHGRSGPAKTSERIRSKRTSRVGPVPTSRALAIMEPPGSDTEAYVKHSEDSAESDEDKVDNEDSPQWKLLETVKNHLGPSGLPMAEPFWKLPSRRAYPNYYKEIKNPVSLNQIKNKIRRGSYGTLSEVAGDMNIMFENAKQYNAPTSRLYKDSVKLQRLMQQRVQELLDIVQSSSSDDESLSSVKNQTQVQTPRPRGRPRINPLPAPAPPPAAAPLVLKPNMPLKKKLHYVAKQLVEFTCSDGRQPMLLFMEKPSKKLYPEYYNVIDRPIDMLTIEANIKNDRYNSLEEMIADFRLMFSNCRHFNEEGSMVYEDANLLERVMNEKIKELNSNYEKKTPMKSLKAQPRSRQLSPFEQKLRTLYDAIRDYRDPKANRQLALIFMKLPSKTEYPDYYELIKNPIDMEKIAHKLKSNSYSSVNELASDFILMFDNACKYNEPDSQIYKDALILHRVCLQTKQMLSQDDDAVPDVPAAVQELLLNLFTTVYNHQDEEGRCYSDSMAELPEHDETNNGEKMRAISLDLVKRRLDKGLYKRLDHFQQDMFAVFERARRLSRTDSQIFEDSVELQCYYIEQRDALCRGTLQSPALTFTRDTVSTSVELIKQCKLLQENDDEDETRSSTDDSMPSGDTNLQSQYRKGDFLYIQPEKGNKDFNIVQVERLWTNSDGITMLYCNMYFRPQETFHIRTRKFLQQEVFKTDVYKPVPLDRVVGPCYVMNVKEYFKFRPEGYADKDVYVCESRYSTKHRWFKKIRAWEGSEKEVSIVPREVALEPQRTVSVFRERVEKHKDELAELEVLENVQEKERPDVVMYNPLGTDDENTYYEQYNTVCSGVIKTGDYVYVVTDGGKQMIAQVDTIWETGDNKCYFRGPFLIFPAEVLNIINKPFYKQEVLLTTIHDTSPLVGIVGKCSVLDYDDYLKCRPTEISEGDVYVCESIYDESNRVARKLKSGLRKFEHTKDVTVDEIYFFPKPLGPPALASTHDVQSASTAFAQKPQHQTMHMDPTDCKPQFTNLINTTIGSQDVEMILENSLDDSSLASPATPLSTGGNSNPYNPSLSVTPSQERIASQTATPVTGKKKKEQKQKIVTGYILYSSEVRKAVIANNPEATFGDISRIVGNEWRSLPAATKQSWEERAARCNEETAARLADEMRELAQHTTMEMTYECAWDTCDCQFEDLTDCMEHCIGDGGNTGHIQQHYRGSFSEYPCLWRNCARVRKGQAPFPNLPRLLRHVRDLHVNKGNGRLMAVHERSRNFMLSSKKPRPAVRSGVMSPGGTNSSSNSQASLSGMSPMARNTPSPGAGEGTSTTIPTAPPVRAGLDPLFVTAPPRAQRVTHSEAYIRYIEGLHSEQKYITPWEKSLTPMPANPDPAQFNMQKIPGHWITDEAINGYLTHDKNLAEADIQKMDQNQKVLKGLCSLRDFMMKDALCLYKNLQINGF
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Uniprot
A0A0N1IA99
A0A194QDY4
A0A1E1WD41
A0A1Y1MQU5
A0A347ZJ98
D6W9A6
+ More
A0A2J7PNZ9 A0A2J7PP20 A0A347ZJ97 A0A139WMA9 A0A139WMV1 A0A2J7PNZ0 A0A2J7PNZ6 A0A2J7PP34 F4X485 A0A151I4A6 A0A151JUZ6 A0A158N9H8 A0A195E1N3 A0A087ZVN9 A0A2A3E8C5 A0A151XGH2 A0A026WDM8 E2BFH5 E2AE67 A0A347ZJ99 A0A154P2J9 A0A1B6CN38 A0A1B6DBT0 W4VRH5 N6TCL8 B0WJC3 A0A1Q3F0S5 A0A1Q3F0Q7 A0A1Q3F0R7 A0A069DY56 Q17Q18 A0A1B0CHW6 A0A182H455 E0VJE8 A0A182FSM1 A0A1B6D864 A0A084WT55 A0A182K626 A0A2M4A8V7 A0A2M4A933 A0A182UT86 W5JGM0 A0A182NR38 A0A182R530 A0A182MT35 T1IDD9 Q7PWX5 A0A1A9UFG6 A0A1A9YQ25 A0A1A9ZT14 A0A1B0G2F8 A0A2M4B8P3 A0A0P4Y990 A0A0P6H6V1 A0A164RK23 A0A2M4B8S1 A0A1A9WML7 A0A182Q1C5 A0A1B0B4X1 A0A182VWB5 A0A182ISH9 A0A0N8C3Z9 B4LWE2 B4K5I8 A0A0P6AXD5 A0A232F4D5 B4NGP4 A0A0N8CQQ3 A0A0L0CBV0 W8BEG0 A0A2M3YZY5 A0A182PR37 A0A2M4A9E2 A0A034VU19 B4JIK7 U4U2H0 A0A1I8MU72 A0A1Y1MVM8 A0A1Y1MQV8 A0A0M4F6I1 A0A1I8PEL3 Q29C06 E9GL93 A0A1W4W1F8 A0A3B0JKD1 B3P6Y7 Q9VC36 A0A0K8WHD7 B4PV44 B7FNJ0 Q7YU13 B3MTV5
A0A2J7PNZ9 A0A2J7PP20 A0A347ZJ97 A0A139WMA9 A0A139WMV1 A0A2J7PNZ0 A0A2J7PNZ6 A0A2J7PP34 F4X485 A0A151I4A6 A0A151JUZ6 A0A158N9H8 A0A195E1N3 A0A087ZVN9 A0A2A3E8C5 A0A151XGH2 A0A026WDM8 E2BFH5 E2AE67 A0A347ZJ99 A0A154P2J9 A0A1B6CN38 A0A1B6DBT0 W4VRH5 N6TCL8 B0WJC3 A0A1Q3F0S5 A0A1Q3F0Q7 A0A1Q3F0R7 A0A069DY56 Q17Q18 A0A1B0CHW6 A0A182H455 E0VJE8 A0A182FSM1 A0A1B6D864 A0A084WT55 A0A182K626 A0A2M4A8V7 A0A2M4A933 A0A182UT86 W5JGM0 A0A182NR38 A0A182R530 A0A182MT35 T1IDD9 Q7PWX5 A0A1A9UFG6 A0A1A9YQ25 A0A1A9ZT14 A0A1B0G2F8 A0A2M4B8P3 A0A0P4Y990 A0A0P6H6V1 A0A164RK23 A0A2M4B8S1 A0A1A9WML7 A0A182Q1C5 A0A1B0B4X1 A0A182VWB5 A0A182ISH9 A0A0N8C3Z9 B4LWE2 B4K5I8 A0A0P6AXD5 A0A232F4D5 B4NGP4 A0A0N8CQQ3 A0A0L0CBV0 W8BEG0 A0A2M3YZY5 A0A182PR37 A0A2M4A9E2 A0A034VU19 B4JIK7 U4U2H0 A0A1I8MU72 A0A1Y1MVM8 A0A1Y1MQV8 A0A0M4F6I1 A0A1I8PEL3 Q29C06 E9GL93 A0A1W4W1F8 A0A3B0JKD1 B3P6Y7 Q9VC36 A0A0K8WHD7 B4PV44 B7FNJ0 Q7YU13 B3MTV5
Pubmed
26354079
28004739
18362917
19820115
21719571
21347285
+ More
24508170 20798317 23537049 26334808 17510324 26483478 20566863 24438588 20920257 23761445 12364791 17994087 28648823 26108605 24495485 25348373 25315136 15632085 21292972 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304
24508170 20798317 23537049 26334808 17510324 26483478 20566863 24438588 20920257 23761445 12364791 17994087 28648823 26108605 24495485 25348373 25315136 15632085 21292972 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304
EMBL
KQ459941
KPJ19149.1
KQ459193
KPJ03160.1
GDQN01006154
JAT84900.1
+ More
GEZM01024033 JAV88043.1 FX985666 BBA84404.1 KQ971312 EEZ98189.2 NEVH01023281 PNF18062.1 PNF18061.1 FX985665 BBA84403.1 KYB29178.1 KYB29177.1 PNF18060.1 PNF18063.1 PNF18059.1 GL888628 EGI58753.1 KQ976461 KYM84717.1 KQ981713 KYN37353.1 ADTU01009654 KQ979814 KYN19006.1 KZ288327 PBC27958.1 KQ982169 KYQ59457.1 KK107260 EZA54195.1 GL448037 EFN85497.1 GL438827 EFN68300.1 FX985667 BBA84405.1 KQ434808 KZC06165.1 GEDC01022543 JAS14755.1 GEDC01014233 JAS23065.1 GANO01003015 JAB56856.1 APGK01035444 APGK01035445 KB740925 ENN78044.1 DS231958 EDS29058.1 GFDL01013888 JAV21157.1 GFDL01013905 JAV21140.1 GFDL01013904 JAV21141.1 GBGD01000064 JAC88825.1 CH477188 EAT48776.1 AJWK01012809 JXUM01025365 JXUM01025366 JXUM01025367 KQ560720 KXJ81153.1 DS235222 EEB13504.1 GEDC01015437 JAS21861.1 ATLV01026856 KE525420 KFB53399.1 GGFK01003839 MBW37160.1 GGFK01003807 MBW37128.1 ADMH02001470 ETN62468.1 AXCM01001187 ACPB03010353 AAAB01008987 EAA01211.4 CCAG010016757 GGFJ01000265 MBW49406.1 GDIP01231710 JAI91691.1 GDIQ01024407 JAN70330.1 LRGB01002190 KZS08730.1 GGFJ01000259 MBW49400.1 AXCN02001477 JXJN01008572 GDIQ01117067 JAL34659.1 CH940650 EDW66579.1 CH933806 EDW14025.1 GDIP01022954 JAM80761.1 NNAY01001049 OXU25338.1 CH964272 EDW84391.1 GDIP01108223 JAL95491.1 JRES01000634 KNC29730.1 GAMC01009508 JAB97047.1 GGFM01001076 MBW21827.1 GGFK01004086 MBW37407.1 GAKP01013632 JAC45320.1 CH916369 EDV93088.1 KB631924 ERL87262.1 GEZM01024031 JAV88046.1 GEZM01024032 JAV88044.1 CP012526 ALC47662.1 CM000070 EAL26840.2 GL732550 EFX79816.1 OUUW01000005 SPP81263.1 CH954182 EDV53807.1 AE014297 AAF56339.1 GDHF01001746 JAI50568.1 CM000160 EDW98893.1 BT053680 ACK77595.1 BT010048 AAQ22517.1 CH902623 EDV30236.1
GEZM01024033 JAV88043.1 FX985666 BBA84404.1 KQ971312 EEZ98189.2 NEVH01023281 PNF18062.1 PNF18061.1 FX985665 BBA84403.1 KYB29178.1 KYB29177.1 PNF18060.1 PNF18063.1 PNF18059.1 GL888628 EGI58753.1 KQ976461 KYM84717.1 KQ981713 KYN37353.1 ADTU01009654 KQ979814 KYN19006.1 KZ288327 PBC27958.1 KQ982169 KYQ59457.1 KK107260 EZA54195.1 GL448037 EFN85497.1 GL438827 EFN68300.1 FX985667 BBA84405.1 KQ434808 KZC06165.1 GEDC01022543 JAS14755.1 GEDC01014233 JAS23065.1 GANO01003015 JAB56856.1 APGK01035444 APGK01035445 KB740925 ENN78044.1 DS231958 EDS29058.1 GFDL01013888 JAV21157.1 GFDL01013905 JAV21140.1 GFDL01013904 JAV21141.1 GBGD01000064 JAC88825.1 CH477188 EAT48776.1 AJWK01012809 JXUM01025365 JXUM01025366 JXUM01025367 KQ560720 KXJ81153.1 DS235222 EEB13504.1 GEDC01015437 JAS21861.1 ATLV01026856 KE525420 KFB53399.1 GGFK01003839 MBW37160.1 GGFK01003807 MBW37128.1 ADMH02001470 ETN62468.1 AXCM01001187 ACPB03010353 AAAB01008987 EAA01211.4 CCAG010016757 GGFJ01000265 MBW49406.1 GDIP01231710 JAI91691.1 GDIQ01024407 JAN70330.1 LRGB01002190 KZS08730.1 GGFJ01000259 MBW49400.1 AXCN02001477 JXJN01008572 GDIQ01117067 JAL34659.1 CH940650 EDW66579.1 CH933806 EDW14025.1 GDIP01022954 JAM80761.1 NNAY01001049 OXU25338.1 CH964272 EDW84391.1 GDIP01108223 JAL95491.1 JRES01000634 KNC29730.1 GAMC01009508 JAB97047.1 GGFM01001076 MBW21827.1 GGFK01004086 MBW37407.1 GAKP01013632 JAC45320.1 CH916369 EDV93088.1 KB631924 ERL87262.1 GEZM01024031 JAV88046.1 GEZM01024032 JAV88044.1 CP012526 ALC47662.1 CM000070 EAL26840.2 GL732550 EFX79816.1 OUUW01000005 SPP81263.1 CH954182 EDV53807.1 AE014297 AAF56339.1 GDHF01001746 JAI50568.1 CM000160 EDW98893.1 BT053680 ACK77595.1 BT010048 AAQ22517.1 CH902623 EDV30236.1
Proteomes
UP000053240
UP000053268
UP000007266
UP000235965
UP000007755
UP000078540
+ More
UP000078541 UP000005205 UP000078492 UP000005203 UP000242457 UP000075809 UP000053097 UP000008237 UP000000311 UP000076502 UP000019118 UP000002320 UP000008820 UP000092461 UP000069940 UP000249989 UP000009046 UP000069272 UP000030765 UP000075881 UP000075903 UP000000673 UP000075884 UP000075900 UP000075883 UP000015103 UP000007062 UP000078200 UP000092443 UP000092445 UP000092444 UP000076858 UP000091820 UP000075886 UP000092460 UP000075920 UP000075880 UP000008792 UP000009192 UP000215335 UP000007798 UP000037069 UP000075885 UP000001070 UP000030742 UP000095301 UP000092553 UP000095300 UP000001819 UP000000305 UP000192221 UP000268350 UP000008711 UP000000803 UP000002282 UP000007801
UP000078541 UP000005205 UP000078492 UP000005203 UP000242457 UP000075809 UP000053097 UP000008237 UP000000311 UP000076502 UP000019118 UP000002320 UP000008820 UP000092461 UP000069940 UP000249989 UP000009046 UP000069272 UP000030765 UP000075881 UP000075903 UP000000673 UP000075884 UP000075900 UP000075883 UP000015103 UP000007062 UP000078200 UP000092443 UP000092445 UP000092444 UP000076858 UP000091820 UP000075886 UP000092460 UP000075920 UP000075880 UP000008792 UP000009192 UP000215335 UP000007798 UP000037069 UP000075885 UP000001070 UP000030742 UP000095301 UP000092553 UP000095300 UP000001819 UP000000305 UP000192221 UP000268350 UP000008711 UP000000803 UP000002282 UP000007801
Pfam
Interpro
IPR037382
Rsc/polybromo
+ More
IPR009071 HMG_box_dom
IPR018359 Bromodomain_CS
IPR001487 Bromodomain
IPR001025 BAH_dom
IPR036427 Bromodomain-like_sf
IPR027180 RSC1/2/4
IPR037968 PBRM1_BD5
IPR036910 HMG_box_dom_sf
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR036291 NAD(P)-bd_dom_sf
IPR002225 3Beta_OHSteriod_DH/Estase
IPR013087 Znf_C2H2_type
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR029058 AB_hydrolase
IPR009071 HMG_box_dom
IPR018359 Bromodomain_CS
IPR001487 Bromodomain
IPR001025 BAH_dom
IPR036427 Bromodomain-like_sf
IPR027180 RSC1/2/4
IPR037968 PBRM1_BD5
IPR036910 HMG_box_dom_sf
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR036291 NAD(P)-bd_dom_sf
IPR002225 3Beta_OHSteriod_DH/Estase
IPR013087 Znf_C2H2_type
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR029058 AB_hydrolase
Gene 3D
ProteinModelPortal
A0A0N1IA99
A0A194QDY4
A0A1E1WD41
A0A1Y1MQU5
A0A347ZJ98
D6W9A6
+ More
A0A2J7PNZ9 A0A2J7PP20 A0A347ZJ97 A0A139WMA9 A0A139WMV1 A0A2J7PNZ0 A0A2J7PNZ6 A0A2J7PP34 F4X485 A0A151I4A6 A0A151JUZ6 A0A158N9H8 A0A195E1N3 A0A087ZVN9 A0A2A3E8C5 A0A151XGH2 A0A026WDM8 E2BFH5 E2AE67 A0A347ZJ99 A0A154P2J9 A0A1B6CN38 A0A1B6DBT0 W4VRH5 N6TCL8 B0WJC3 A0A1Q3F0S5 A0A1Q3F0Q7 A0A1Q3F0R7 A0A069DY56 Q17Q18 A0A1B0CHW6 A0A182H455 E0VJE8 A0A182FSM1 A0A1B6D864 A0A084WT55 A0A182K626 A0A2M4A8V7 A0A2M4A933 A0A182UT86 W5JGM0 A0A182NR38 A0A182R530 A0A182MT35 T1IDD9 Q7PWX5 A0A1A9UFG6 A0A1A9YQ25 A0A1A9ZT14 A0A1B0G2F8 A0A2M4B8P3 A0A0P4Y990 A0A0P6H6V1 A0A164RK23 A0A2M4B8S1 A0A1A9WML7 A0A182Q1C5 A0A1B0B4X1 A0A182VWB5 A0A182ISH9 A0A0N8C3Z9 B4LWE2 B4K5I8 A0A0P6AXD5 A0A232F4D5 B4NGP4 A0A0N8CQQ3 A0A0L0CBV0 W8BEG0 A0A2M3YZY5 A0A182PR37 A0A2M4A9E2 A0A034VU19 B4JIK7 U4U2H0 A0A1I8MU72 A0A1Y1MVM8 A0A1Y1MQV8 A0A0M4F6I1 A0A1I8PEL3 Q29C06 E9GL93 A0A1W4W1F8 A0A3B0JKD1 B3P6Y7 Q9VC36 A0A0K8WHD7 B4PV44 B7FNJ0 Q7YU13 B3MTV5
A0A2J7PNZ9 A0A2J7PP20 A0A347ZJ97 A0A139WMA9 A0A139WMV1 A0A2J7PNZ0 A0A2J7PNZ6 A0A2J7PP34 F4X485 A0A151I4A6 A0A151JUZ6 A0A158N9H8 A0A195E1N3 A0A087ZVN9 A0A2A3E8C5 A0A151XGH2 A0A026WDM8 E2BFH5 E2AE67 A0A347ZJ99 A0A154P2J9 A0A1B6CN38 A0A1B6DBT0 W4VRH5 N6TCL8 B0WJC3 A0A1Q3F0S5 A0A1Q3F0Q7 A0A1Q3F0R7 A0A069DY56 Q17Q18 A0A1B0CHW6 A0A182H455 E0VJE8 A0A182FSM1 A0A1B6D864 A0A084WT55 A0A182K626 A0A2M4A8V7 A0A2M4A933 A0A182UT86 W5JGM0 A0A182NR38 A0A182R530 A0A182MT35 T1IDD9 Q7PWX5 A0A1A9UFG6 A0A1A9YQ25 A0A1A9ZT14 A0A1B0G2F8 A0A2M4B8P3 A0A0P4Y990 A0A0P6H6V1 A0A164RK23 A0A2M4B8S1 A0A1A9WML7 A0A182Q1C5 A0A1B0B4X1 A0A182VWB5 A0A182ISH9 A0A0N8C3Z9 B4LWE2 B4K5I8 A0A0P6AXD5 A0A232F4D5 B4NGP4 A0A0N8CQQ3 A0A0L0CBV0 W8BEG0 A0A2M3YZY5 A0A182PR37 A0A2M4A9E2 A0A034VU19 B4JIK7 U4U2H0 A0A1I8MU72 A0A1Y1MVM8 A0A1Y1MQV8 A0A0M4F6I1 A0A1I8PEL3 Q29C06 E9GL93 A0A1W4W1F8 A0A3B0JKD1 B3P6Y7 Q9VC36 A0A0K8WHD7 B4PV44 B7FNJ0 Q7YU13 B3MTV5
PDB
1W4S
E-value=8.45655e-38,
Score=400
Ontologies
GO
GO:0003682
GO:0006338
GO:0003677
GO:0016586
GO:0043044
GO:0006368
GO:0006337
GO:0016021
GO:0004930
GO:0007166
GO:0006694
GO:0003854
GO:0016787
GO:0007305
GO:0031936
GO:0035060
GO:0007306
GO:0007480
GO:0000122
GO:0005700
GO:0005515
GO:0007608
GO:0016020
GO:0020037
GO:0030117
GO:0006418
GO:0003707
GO:0006281
GO:0006259
PANTHER
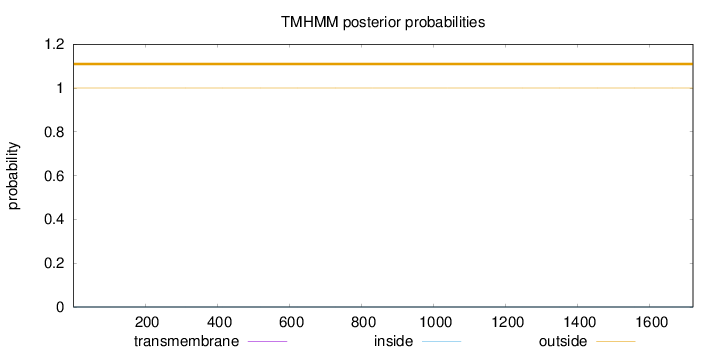
Topology
Length:
1720
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1720
Population Genetic Test Statistics
Pi
262.249532
Theta
17.259729
Tajima's D
1.511861
CLR
0.366528
CSRT
0.797210139493025
Interpretation
Uncertain
