Gene
KWMTBOMO15624
Pre Gene Modal
BGIBMGA010844
Annotation
PREDICTED:_odorant_receptor_4-like_[Plutella_xylostella]
Full name
Odorant receptor
+ More
Odorant receptor 4
Odorant receptor 4
Location in the cell
PlasmaMembrane Reliability : 4.863
Sequence
CDS
ATGGCCAAATCCTACAAGATAGTGTATTATCGTCCGATCTATGAGAATTTGATTTATGAGCTACGGGATATGTGGCCGAAGGGCTGCGTTGCCGACGAAGAACATGCAATCATCTGTACAGCCCTCAAACAACTCAATTATGTCGTGCAGGTATTTATAATATGGAGCTTATTTAGGTTACTACTGGTGCACAACGCCCTACTAGTAATATTCCTGTCGCCGCCATTCGTGGAGATCATCAAACAGGTCGCTGGTTGGGACGTTCCTCTGATCCTGCCCTTCTTCTACTGGTTTCCCTTCGACCCGTTCCAGCGAATCTACTACGAAACGATTCTTATCTTTCAAACTTGGCACGGGCTAATTACAATCTGGTTTATGCTGTGCGGAGATTTACTGTTCTGTATATTCTTGAGCCATATTTCGACGCAGTTCGACTTACTGGCAGTGAGGATCAAGAGAATGGTATATGTGCCTGTTGATAAACAGCTAATTGAAGAATACCCTTTGGGTGAGTATCAACGGAGCTAA
Protein
MAKSYKIVYYRPIYENLIYELRDMWPKGCVADEEHAIICTALKQLNYVVQVFIIWSLFRLLLVHNALLVIFLSPPFVEIIKQVAGWDVPLILPFFYWFPFDPFQRIYYETILIFQTWHGLITIWFMLCGDLLFCIFLSHISTQFDLLAVRIKRMVYVPVDKQLIEEYPLGEYQRS
Summary
Description
Odorant receptor that specifically recognizes the human odorant sulcatone (6-methylhept-5-en-2-onesul), a volatile odorant emitted at uniquely high levels in humans, thereby playing a key role in mosquito's preference in biting human compared to other animals.
Miscellaneous
Expressed at much higher levels in mosquito strains that preferentially bite human compared to 'ancestral' strains that prefer to bite non-human animals.
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Belongs to the heteromeric odorant receptor channel (TC 1.A.69) family.
Belongs to the transient receptor (TC 1.A.4) family.
Belongs to the heteromeric odorant receptor channel (TC 1.A.69) family.
Belongs to the transient receptor (TC 1.A.4) family.
Keywords
Complete proteome
Membrane
Olfaction
Polymorphism
Receptor
Reference proteome
Sensory transduction
Transducer
Transmembrane
Transmembrane helix
Feature
chain Odorant receptor 4
Uniprot
A0A2H1WMI9
A0A3G6V6Q3
A0A2A4JF91
B7S6U6
A0A2H1V8L9
M1R1W5
+ More
A0A194QC90 A0A2L1TG47 A0A0E4B5A7 A0A1V1WCD1 H9JU52 A0A2W1BU51 A0A2A4K396 A0A076E7J5 A0A0P1J1P1 A0A386H997 A0A2K8GKS1 A0A2K8GL45 A0A076E9D1 I3Y3I6 A0A212FA57 A0A3Q9NC10 A0A1Y9TJI5 A0A1S5XXQ9 A0A223HDC2 A0A0V0J0Y1 A0A1B3B724 A0A146JW29 A0A0S1TRB5 A0A0L7LCD2 A0A1B2G2N8 A0A1X9PB26 A0A212ELH4 A0A1B0DM33 Q16NV7 A0A1B0GLR6 A0A1S7UE45 Q16EI9 A0A2K7P6E1 A0A0E4B5I0 A0A386H919 A0A097IYJ9 A0A097ITW8 A0A146JVM3 A0A0B5CMU9 A0A0B5GEY2 A0A0P1IW06 A0A0P1IVT0 A0A2A4K768 A0A0N7MCA4 A0A0K8TVB7 A0A3F2ZDH2 A0A2H1WTJ9 A0A182YJS6 A0A1L2BLA8 H9A5N1 A0A182VQZ4 A0A0N7MCA7 A0A0K8TUR8 A0A182R1K2 A0A076EDF5 A0A386H9C0 A0A0B5GI99 A0A0B5CN07 A0A223HDG1 A0A2K8GKS4 A0A182MBR6 Q6A1K2 A0A3G6V7H7 A0A1S5XXQ5 A0A0P1IW02 A0A076E621 A0A1B3B759 A0A076EDH7 A0A076E9C3 A0A182FQJ2 Q16MN0 A0A1S4FVR6 A0A2A4K114 A0A2A4K9L1 A0A075T619 R4JUT8
A0A194QC90 A0A2L1TG47 A0A0E4B5A7 A0A1V1WCD1 H9JU52 A0A2W1BU51 A0A2A4K396 A0A076E7J5 A0A0P1J1P1 A0A386H997 A0A2K8GKS1 A0A2K8GL45 A0A076E9D1 I3Y3I6 A0A212FA57 A0A3Q9NC10 A0A1Y9TJI5 A0A1S5XXQ9 A0A223HDC2 A0A0V0J0Y1 A0A1B3B724 A0A146JW29 A0A0S1TRB5 A0A0L7LCD2 A0A1B2G2N8 A0A1X9PB26 A0A212ELH4 A0A1B0DM33 Q16NV7 A0A1B0GLR6 A0A1S7UE45 Q16EI9 A0A2K7P6E1 A0A0E4B5I0 A0A386H919 A0A097IYJ9 A0A097ITW8 A0A146JVM3 A0A0B5CMU9 A0A0B5GEY2 A0A0P1IW06 A0A0P1IVT0 A0A2A4K768 A0A0N7MCA4 A0A0K8TVB7 A0A3F2ZDH2 A0A2H1WTJ9 A0A182YJS6 A0A1L2BLA8 H9A5N1 A0A182VQZ4 A0A0N7MCA7 A0A0K8TUR8 A0A182R1K2 A0A076EDF5 A0A386H9C0 A0A0B5GI99 A0A0B5CN07 A0A223HDG1 A0A2K8GKS4 A0A182MBR6 Q6A1K2 A0A3G6V7H7 A0A1S5XXQ5 A0A0P1IW02 A0A076E621 A0A1B3B759 A0A076EDH7 A0A076E9C3 A0A182FQJ2 Q16MN0 A0A1S4FVR6 A0A2A4K114 A0A2A4K9L1 A0A075T619 R4JUT8
Pubmed
EMBL
ODYU01009683
SOQ54285.1
MH193978
AZB49424.1
NWSH01001752
PCG70244.1
+ More
EF396285 ABQ84982.1 ODYU01001242 SOQ37185.1 JX999588 AGG08878.1 KQ459193 KPJ03158.1 KY707183 AVF19625.1 LC002728 BAR43476.1 GENK01000082 JAV45831.1 BABH01004087 BABH01004088 KZ149935 PZC77164.1 NWSH01000176 PCG78731.1 KF487664 AII01062.1 LN885129 CUQ99418.1 MG546600 AYD42223.1 KX585336 ARO70229.1 KY225492 ARO70504.1 KF487711 AII01109.1 JQ794816 AFL70821.1 AGBW02009516 OWR50610.1 MH050653 AZT78919.1 KX084480 ARO76435.1 KY399306 AQQ73537.1 KY283727 AST36386.1 GDKB01000059 JAP38437.1 KT588111 AOE48021.1 GEDO01000039 JAP88587.1 KR935708 ALM26200.1 JTDY01001727 KOB73069.1 KU598903 ANZ03149.1 KX084484 ARO76439.1 AGBW02014048 OWR42350.1 AJVK01036856 AJVK01036857 BK006141 CH477806 DAA80356.1 EAT36037.2 AJWK01030596 BK006140 DAA80355.1 KF801614 KF801615 KF801617 KF801618 KF801619 KF801620 KF801621 CH478696 EAT32646.1 LC002721 BAR43469.1 MG546625 AYD42248.1 KM655564 AIT72013.1 KM597226 AIT69903.1 KX084477 GEDO01000063 ARO76432.1 JAP88563.1 KM678326 AJE25899.1 KM892400 AJF23815.1 LN885103 CUQ99392.1 LN885127 CUQ99416.1 NWSH01000058 PCG80087.1 LN885107 CUQ99396.1 GCVX01000215 JAI18015.1 AJWK01027546 ODYU01010558 SOQ55754.1 KT860047 ALS03874.1 JN836681 AFC91721.1 LN885132 CUQ99421.1 GCVX01000214 JAI18016.1 KF487677 AII01075.1 MG546642 AYD42265.1 KM892401 AJF23816.1 KM678327 AJE25900.1 KY283749 AST36408.1 KX585344 ARO70237.1 AXCM01002463 AJ748327 CAG38113.1 MH193980 AZB49426.1 KY399305 AQQ73536.1 LN885105 CUQ99394.1 KF487703 AII01101.1 KT588139 AOE48049.1 KF487702 AII01100.1 KF487701 AII01099.1 BK006143 CH477856 DAA80358.1 EAT35587.2 NWSH01000302 PCG77608.1 NWSH01000040 PCG80352.1 KF768661 AIG51855.1 JX982526 AGK90001.1
EF396285 ABQ84982.1 ODYU01001242 SOQ37185.1 JX999588 AGG08878.1 KQ459193 KPJ03158.1 KY707183 AVF19625.1 LC002728 BAR43476.1 GENK01000082 JAV45831.1 BABH01004087 BABH01004088 KZ149935 PZC77164.1 NWSH01000176 PCG78731.1 KF487664 AII01062.1 LN885129 CUQ99418.1 MG546600 AYD42223.1 KX585336 ARO70229.1 KY225492 ARO70504.1 KF487711 AII01109.1 JQ794816 AFL70821.1 AGBW02009516 OWR50610.1 MH050653 AZT78919.1 KX084480 ARO76435.1 KY399306 AQQ73537.1 KY283727 AST36386.1 GDKB01000059 JAP38437.1 KT588111 AOE48021.1 GEDO01000039 JAP88587.1 KR935708 ALM26200.1 JTDY01001727 KOB73069.1 KU598903 ANZ03149.1 KX084484 ARO76439.1 AGBW02014048 OWR42350.1 AJVK01036856 AJVK01036857 BK006141 CH477806 DAA80356.1 EAT36037.2 AJWK01030596 BK006140 DAA80355.1 KF801614 KF801615 KF801617 KF801618 KF801619 KF801620 KF801621 CH478696 EAT32646.1 LC002721 BAR43469.1 MG546625 AYD42248.1 KM655564 AIT72013.1 KM597226 AIT69903.1 KX084477 GEDO01000063 ARO76432.1 JAP88563.1 KM678326 AJE25899.1 KM892400 AJF23815.1 LN885103 CUQ99392.1 LN885127 CUQ99416.1 NWSH01000058 PCG80087.1 LN885107 CUQ99396.1 GCVX01000215 JAI18015.1 AJWK01027546 ODYU01010558 SOQ55754.1 KT860047 ALS03874.1 JN836681 AFC91721.1 LN885132 CUQ99421.1 GCVX01000214 JAI18016.1 KF487677 AII01075.1 MG546642 AYD42265.1 KM892401 AJF23816.1 KM678327 AJE25900.1 KY283749 AST36408.1 KX585344 ARO70237.1 AXCM01002463 AJ748327 CAG38113.1 MH193980 AZB49426.1 KY399305 AQQ73536.1 LN885105 CUQ99394.1 KF487703 AII01101.1 KT588139 AOE48049.1 KF487702 AII01100.1 KF487701 AII01099.1 BK006143 CH477856 DAA80358.1 EAT35587.2 NWSH01000302 PCG77608.1 NWSH01000040 PCG80352.1 KF768661 AIG51855.1 JX982526 AGK90001.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
A0A2H1WMI9
A0A3G6V6Q3
A0A2A4JF91
B7S6U6
A0A2H1V8L9
M1R1W5
+ More
A0A194QC90 A0A2L1TG47 A0A0E4B5A7 A0A1V1WCD1 H9JU52 A0A2W1BU51 A0A2A4K396 A0A076E7J5 A0A0P1J1P1 A0A386H997 A0A2K8GKS1 A0A2K8GL45 A0A076E9D1 I3Y3I6 A0A212FA57 A0A3Q9NC10 A0A1Y9TJI5 A0A1S5XXQ9 A0A223HDC2 A0A0V0J0Y1 A0A1B3B724 A0A146JW29 A0A0S1TRB5 A0A0L7LCD2 A0A1B2G2N8 A0A1X9PB26 A0A212ELH4 A0A1B0DM33 Q16NV7 A0A1B0GLR6 A0A1S7UE45 Q16EI9 A0A2K7P6E1 A0A0E4B5I0 A0A386H919 A0A097IYJ9 A0A097ITW8 A0A146JVM3 A0A0B5CMU9 A0A0B5GEY2 A0A0P1IW06 A0A0P1IVT0 A0A2A4K768 A0A0N7MCA4 A0A0K8TVB7 A0A3F2ZDH2 A0A2H1WTJ9 A0A182YJS6 A0A1L2BLA8 H9A5N1 A0A182VQZ4 A0A0N7MCA7 A0A0K8TUR8 A0A182R1K2 A0A076EDF5 A0A386H9C0 A0A0B5GI99 A0A0B5CN07 A0A223HDG1 A0A2K8GKS4 A0A182MBR6 Q6A1K2 A0A3G6V7H7 A0A1S5XXQ5 A0A0P1IW02 A0A076E621 A0A1B3B759 A0A076EDH7 A0A076E9C3 A0A182FQJ2 Q16MN0 A0A1S4FVR6 A0A2A4K114 A0A2A4K9L1 A0A075T619 R4JUT8
A0A194QC90 A0A2L1TG47 A0A0E4B5A7 A0A1V1WCD1 H9JU52 A0A2W1BU51 A0A2A4K396 A0A076E7J5 A0A0P1J1P1 A0A386H997 A0A2K8GKS1 A0A2K8GL45 A0A076E9D1 I3Y3I6 A0A212FA57 A0A3Q9NC10 A0A1Y9TJI5 A0A1S5XXQ9 A0A223HDC2 A0A0V0J0Y1 A0A1B3B724 A0A146JW29 A0A0S1TRB5 A0A0L7LCD2 A0A1B2G2N8 A0A1X9PB26 A0A212ELH4 A0A1B0DM33 Q16NV7 A0A1B0GLR6 A0A1S7UE45 Q16EI9 A0A2K7P6E1 A0A0E4B5I0 A0A386H919 A0A097IYJ9 A0A097ITW8 A0A146JVM3 A0A0B5CMU9 A0A0B5GEY2 A0A0P1IW06 A0A0P1IVT0 A0A2A4K768 A0A0N7MCA4 A0A0K8TVB7 A0A3F2ZDH2 A0A2H1WTJ9 A0A182YJS6 A0A1L2BLA8 H9A5N1 A0A182VQZ4 A0A0N7MCA7 A0A0K8TUR8 A0A182R1K2 A0A076EDF5 A0A386H9C0 A0A0B5GI99 A0A0B5CN07 A0A223HDG1 A0A2K8GKS4 A0A182MBR6 Q6A1K2 A0A3G6V7H7 A0A1S5XXQ5 A0A0P1IW02 A0A076E621 A0A1B3B759 A0A076EDH7 A0A076E9C3 A0A182FQJ2 Q16MN0 A0A1S4FVR6 A0A2A4K114 A0A2A4K9L1 A0A075T619 R4JUT8
Ontologies
GO
Topology
Subcellular location
Cell membrane
Membrane
Membrane
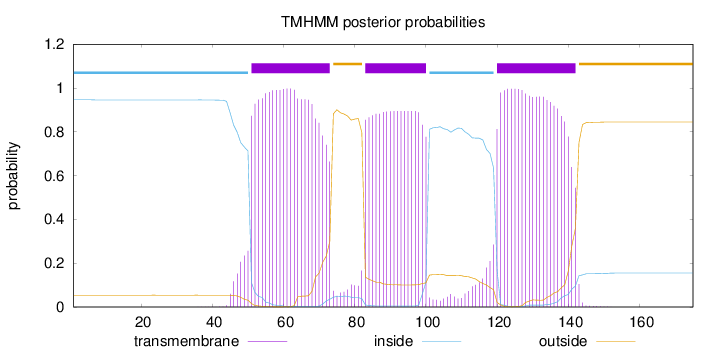
Length:
175
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
61.5911
Exp number, first 60 AAs:
10.66653
Total prob of N-in:
0.94603
POSSIBLE N-term signal
sequence
inside
1 - 50
TMhelix
51 - 73
outside
74 - 82
TMhelix
83 - 100
inside
101 - 119
TMhelix
120 - 142
outside
143 - 175
Population Genetic Test Statistics
Pi
198.909469
Theta
159.813285
Tajima's D
0.702966
CLR
0
CSRT
0.572921353932303
Interpretation
Uncertain
