Gene
KWMTBOMO15623
Pre Gene Modal
BGIBMGA010713
Annotation
PREDICTED:_soluble_guanylate_cyclase_88E_isoform_X1_[Bombyx_mori]
Full name
Soluble guanylate cyclase 88E
Location in the cell
Nuclear Reliability : 2.947
Sequence
CDS
ATGTACGGGCTTCTACTCGAGAACATGGCGGAGTACATCCGACAGACTTACGGCGAGGAACGCTGGGAGGATATCCGGAGGCAGGCTGGAGTTGAACAGCCTTCATTCTCCGTTCACCAGGTGTACCCGGAGAACATGATCACTAGACTCGCCAAAAAAGCGCAAGAGATGCTCGGCATGTCTGAACGTGAATTCATGGATCAAATGGGCGTTTACTTCGTCGGCTTCGTGTCGCAATACGGCTACGACAGAGTTTTATCAGTGCTGGGACGTCATATGCGAGACTTCCTGAACGGGCTGGATAATCTCCACGAGTACCTGAAGTTCAGTTACCCCAGGATGAGAGCGCCGAGTTTCATTTGCGAAAATGAAACGCGACAAGGCTTGACCTTACATTACAGGTCGAAGAGAAGAGGCTTTGTTTATTACGCTATGGGCCAGATTCGGGAGGTAGCCCGGCACTTCTACCATAAAGAAATGCGAATAGAGTTGGTCAGGGAAGAGTTATTATTCGACACGGTCCACGTGACCTTTCAACTGACATTCGATAACAGAGCGTTCACTTTAGCGTCTCTCGCGATGACCAGAGAAGAGAAACATCTGCCGATCAGCGCTTCAGTACTCTTCGAGATCTTTCCGTTTTGCATAGTCTTTGGCTCCGACATGGTGGTGAGGAGTATCGGCAATTCGCTAATGGTGATTCTACCAGATCTGGTGGGAAAGAAGATAACGAATTGGTTCGATCTGGTCAGACCTCTAATAGCATTCAAATTTCAAACGATATTAAATCGCACGAACAATATATTCGAGCTGGTAACGGTTGAAGCAGTAATGCACGAGAAGGCACCGGACAAGCGGAACGAGCTGCTTAGACTCTCCGACGAATCTGACGGTACCACTGAAAAGAACCTACGACTTAAAGGTCAAATGATATATATGGATAACTGGCGCATGATGATGTACCTCGGCACTCCTGTGATGCCTGATCTGTCCGCCCTCGTCTCCACAGGACTGTACATCAACGATTTATCTATGCACGACTTCAGCCGAGACTTAATGCTGGCAGGAACGCAACAGTCCGTTGAACTGAAGCTAGCTTTAGACCAAGAACAGCAGAAGAGCAAGAAGCTAGAGGAGTCGATGAGGAAGCTGGACGAAGAAATGAAAAGAACCGATGAACTGCTCTACCAGATGATACCAAAACAGGTCGCTGATAGACTACGGAATGGAGAAAATCCGATAGATACTTGCGAGATGTTCGATAGCGTGTCCATTCTTTTCTCCGACGTCGTCACCTTCACCGAGATCTGCTCCCGGATAACGCCGATGGAGGTGGTCTCAATGCTGAATGCCATGTACTCTATTTTCGACACGCTCACAGAACGCAATAGGGTTTACAAAGTTGAAACTATAGGAGACGCCTACATGGTTGTTTCTGGTGCACCAGAGAAAGAAGACAATCATGCCGAGAAGGTCTGTGATATGGCCCTTGACATGGTCGATGCGATCACAGATTTGAAGGACCCTAGTACTGGTTCTCATTTATCGATAAGAGTAGGAGTGCATTCCGGGGCGGTTGTCGCCGGTATCGTTGGCCTGAAGATGCCTCGCTATTGTCTGTTCGGAGACTCTGTCAACACTGCATCTCGGATGGAATCCACTTCTGAAGCCATGAGGATACACATATCGCAGACCACTCAGGAACTGCTCTCTCCGTCTTACATGGTCACAGAGAGAGGAGAGATACAGGTGAAGGGAAAAGGTGCAATGAAAACCTATTGGCTAGAAGGCCGCGAATCCAGACCCGCTCTCAACAAAGTGATATCACCACAACTTCAGCCGGGAACCGAACTGGAGTGGGAGCGAGCTGCTGACGTCAGAGACAGTATCGCAGAACATTCAGCCCAACAGATGAACAACAAAGACAGCGCACAGGGTTCCGCCAACAGCCAACATTTGGCCAAACCTGTGACGTCAGGACCGAGCTCTTTAAACATCGCAGCTTTAGTGCCGGTTCAGAGCGTGAGGAATAATATGCAGGTGACTACTGTGACGTCACCGGTCGAAGAAAGAAGGATGTATTCTCCGGTAACTTTCCAAGATGTTGCTAGGAGAAGCATCGCTAATTCTCCTAACAGAGCTGATAGGGAGAGAGAGTCGCGATCGAACTCGGCGAGTGTAGCGGGAGGGTGGACAGACGGCGACTCACTCGATCCGATCAAAAATGGGGACAGTCCGGGCTCGCCATTTTGTTCAACCTCACCTTGCAGAGTCGGAACCGCCCCAGCGGTCAAGGAACAGGGGAATGAAGACTTCTCAGAAACGTTCAAAGCTCGAATGTCACCTGCACACTCGGCTCCCGCTACAATGGGAGATTCCCTGTCCACTCTTAGACCAGATACATCTGTTGCCTCGTCGATCGGGCCGCGACAAAGTACTGATGAAATAGAAACGGACACAGAATACCAAGACGCGCACGCGGATTCGAACCAGGCTCACACGTGCGATAGCACGACTGTGCATCCGAAGCCGGGCCGGGTCGGTCGATTCAGAGCTCGGATTGCACCAGGATACCATAGAAATTGTACGGCTCACAAGCAAACCAGCAAAGACAAATCGCACACGGCCCAGGGCGGGAACGTACAGCCACACGGGCATCACCACACGAAGAACGTGAACCACCATCAGTGCTGTGGAGCGTTCGGGAATCCACACGTAAGGCACAAATCAAATAACTCTAGTTGTCACCTAATTTAA
Protein
MYGLLLENMAEYIRQTYGEERWEDIRRQAGVEQPSFSVHQVYPENMITRLAKKAQEMLGMSEREFMDQMGVYFVGFVSQYGYDRVLSVLGRHMRDFLNGLDNLHEYLKFSYPRMRAPSFICENETRQGLTLHYRSKRRGFVYYAMGQIREVARHFYHKEMRIELVREELLFDTVHVTFQLTFDNRAFTLASLAMTREEKHLPISASVLFEIFPFCIVFGSDMVVRSIGNSLMVILPDLVGKKITNWFDLVRPLIAFKFQTILNRTNNIFELVTVEAVMHEKAPDKRNELLRLSDESDGTTEKNLRLKGQMIYMDNWRMMMYLGTPVMPDLSALVSTGLYINDLSMHDFSRDLMLAGTQQSVELKLALDQEQQKSKKLEESMRKLDEEMKRTDELLYQMIPKQVADRLRNGENPIDTCEMFDSVSILFSDVVTFTEICSRITPMEVVSMLNAMYSIFDTLTERNRVYKVETIGDAYMVVSGAPEKEDNHAEKVCDMALDMVDAITDLKDPSTGSHLSIRVGVHSGAVVAGIVGLKMPRYCLFGDSVNTASRMESTSEAMRIHISQTTQELLSPSYMVTERGEIQVKGKGAMKTYWLEGRESRPALNKVISPQLQPGTELEWERAADVRDSIAEHSAQQMNNKDSAQGSANSQHLAKPVTSGPSSLNIAALVPVQSVRNNMQVTTVTSPVEERRMYSPVTFQDVARRSIANSPNRADRERESRSNSASVAGGWTDGDSLDPIKNGDSPGSPFCSTSPCRVGTAPAVKEQGNEDFSETFKARMSPAHSAPATMGDSLSTLRPDTSVASSIGPRQSTDEIETDTEYQDAHADSNQAHTCDSTTVHPKPGRVGRFRARIAPGYHRNCTAHKQTSKDKSHTAQGGNVQPHGHHHTKNVNHHQCCGAFGNPHVRHKSNNSSCHLI
Summary
Description
Heterodimers with Gyc-89Da and Gyc-89Db are activated in response to changing oxygen concentrations, alerting flies to hypoxic environments. Under normal oxygen concentrations, oxygen binds to the heme group and results in low levels of guanylyl cyclase activity. When exposed to reduced oxygen concentrations, the oxygen dissociates from the heme group resulting in activation of the enzyme.
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Cofactor
heme
Subunit
Homodimer, and heterodimer; with Gyc-89Db and Gyc-89Da, in the presence of magnesium or manganese.
Miscellaneous
There are two types of guanylate cyclases: soluble forms and membrane-associated receptor forms.
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Keywords
Alternative splicing
cGMP biosynthesis
Coiled coil
Complete proteome
Cytoplasm
GTP-binding
Heme
Iron
Lyase
Magnesium
Manganese
Metal-binding
Nucleotide-binding
Reference proteome
Feature
chain Soluble guanylate cyclase 88E
splice variant In isoform F.
splice variant In isoform F.
Uniprot
O76340
A0A2H1VA46
A0A212F2T2
A0A2A4IYJ1
H9JMG0
A0A2A4IZH1
+ More
A0A194QCG4 A0A0N1PK87 A0A2J7PCG4 E0V9C1 A0A2J7PCG7 Q5FAN0 A0A0L7RKC4 A0A0L0BWC8 A0A1I8MPR9 A0A0Q9X803 A0A0Q9X9D2 A0A195CRM1 B4KDK3 E2BXW1 A0A154PK07 A0A1A9YAX6 A0A1A9VGU4 A0A026VSM1 F4X5Q2 A0A195BXI0 A0A158NUX0 A0A151IRN9 A0A1A9W1K2 E2B0Y5 A0A1I8P434 A0A1A9ZAA2 A0A151WSX9 J9K9E5 B4NKT4 A0A1I8P3Z3 A0A195FQ75 A0A1B0BM11 K7J2P7 A0A0M8ZVH0 B4HE17 B3P0Q9 B4JV23 Q8INF0 Q8INF0-2 A0A0M3QY21 B4QZ50 A0A1W4WIZ2 A0A1W4WIQ4 B4PS88 A0A1B0FDH5 Q16JH8 A0A182M001 A0A182RPV2 A0A182XVU3 A0A182PKI3 A0A182VYY9 A0A182NS81 A0A182QFQ0 A0A182JIS4 A0A084VHI9 A0A182F6T4 A0A182UMR7 A0A182I080 A0A182XBD8 A0A182KLU5 Q7PYK9 A0A1J1IFM2 A0A310SI97 A0A182TF11 A0A336LTP0 A0A1Y1K6M1 A0A1Y1KEA5 A0A182JZ34 A0A0P6IUP1
A0A194QCG4 A0A0N1PK87 A0A2J7PCG4 E0V9C1 A0A2J7PCG7 Q5FAN0 A0A0L7RKC4 A0A0L0BWC8 A0A1I8MPR9 A0A0Q9X803 A0A0Q9X9D2 A0A195CRM1 B4KDK3 E2BXW1 A0A154PK07 A0A1A9YAX6 A0A1A9VGU4 A0A026VSM1 F4X5Q2 A0A195BXI0 A0A158NUX0 A0A151IRN9 A0A1A9W1K2 E2B0Y5 A0A1I8P434 A0A1A9ZAA2 A0A151WSX9 J9K9E5 B4NKT4 A0A1I8P3Z3 A0A195FQ75 A0A1B0BM11 K7J2P7 A0A0M8ZVH0 B4HE17 B3P0Q9 B4JV23 Q8INF0 Q8INF0-2 A0A0M3QY21 B4QZ50 A0A1W4WIZ2 A0A1W4WIQ4 B4PS88 A0A1B0FDH5 Q16JH8 A0A182M001 A0A182RPV2 A0A182XVU3 A0A182PKI3 A0A182VYY9 A0A182NS81 A0A182QFQ0 A0A182JIS4 A0A084VHI9 A0A182F6T4 A0A182UMR7 A0A182I080 A0A182XBD8 A0A182KLU5 Q7PYK9 A0A1J1IFM2 A0A310SI97 A0A182TF11 A0A336LTP0 A0A1Y1K6M1 A0A1Y1KEA5 A0A182JZ34 A0A0P6IUP1
EC Number
4.6.1.2
Pubmed
EMBL
AF064514
AAD09836.1
ODYU01001452
SOQ37671.1
AGBW02010685
OWR48039.1
+ More
NWSH01004590 PCG64885.1 BABH01016865 PCG64886.1 KQ459193 KPJ03157.1 KQ459941 KPJ19146.1 NEVH01027059 PNF14030.1 DS234991 EEB09977.1 PNF14029.1 AB204559 BAD89804.1 KQ414573 KOC71284.1 JRES01001243 KNC24332.1 CH933806 KRG00662.1 KRG00663.1 KRG00665.1 KRG00664.1 KQ977444 KYN02759.1 EDW13837.2 GL451328 EFN79468.1 KQ434943 KZC12211.1 KK108839 QOIP01000009 EZA46610.1 RLU18620.1 GL888739 EGI58218.1 KQ976401 KYM92651.1 ADTU01026805 KQ981122 KYN09350.1 GL444716 EFN60654.1 KQ982769 KYQ50933.1 ABLF02041086 CH964272 EDW84145.2 KQ981335 KYN42576.1 JXJN01016642 KQ435827 KOX71960.1 CH480815 EDW42109.1 CH954181 EDV48957.1 CH916374 EDV91343.1 AE014297 BT029937 AY376404 AY376405 AAQ84039.1 ABM92811.1 CP012526 ALC46913.1 CM000364 EDX12878.1 CM000160 EDW97508.1 CCAG010010248 CH478008 EAT34434.1 AXCM01002214 AXCN02000202 ATLV01013177 KE524842 KFB37433.1 APCN01002054 AAAB01008987 EAA01162.5 CVRI01000048 CRK99008.1 KQ762687 OAD55536.1 UFQT01000192 SSX21416.1 GEZM01091080 GEZM01091075 JAV57109.1 GEZM01091082 GEZM01091078 JAV57107.1 GDIQ01001129 JAN93608.1
NWSH01004590 PCG64885.1 BABH01016865 PCG64886.1 KQ459193 KPJ03157.1 KQ459941 KPJ19146.1 NEVH01027059 PNF14030.1 DS234991 EEB09977.1 PNF14029.1 AB204559 BAD89804.1 KQ414573 KOC71284.1 JRES01001243 KNC24332.1 CH933806 KRG00662.1 KRG00663.1 KRG00665.1 KRG00664.1 KQ977444 KYN02759.1 EDW13837.2 GL451328 EFN79468.1 KQ434943 KZC12211.1 KK108839 QOIP01000009 EZA46610.1 RLU18620.1 GL888739 EGI58218.1 KQ976401 KYM92651.1 ADTU01026805 KQ981122 KYN09350.1 GL444716 EFN60654.1 KQ982769 KYQ50933.1 ABLF02041086 CH964272 EDW84145.2 KQ981335 KYN42576.1 JXJN01016642 KQ435827 KOX71960.1 CH480815 EDW42109.1 CH954181 EDV48957.1 CH916374 EDV91343.1 AE014297 BT029937 AY376404 AY376405 AAQ84039.1 ABM92811.1 CP012526 ALC46913.1 CM000364 EDX12878.1 CM000160 EDW97508.1 CCAG010010248 CH478008 EAT34434.1 AXCM01002214 AXCN02000202 ATLV01013177 KE524842 KFB37433.1 APCN01002054 AAAB01008987 EAA01162.5 CVRI01000048 CRK99008.1 KQ762687 OAD55536.1 UFQT01000192 SSX21416.1 GEZM01091080 GEZM01091075 JAV57109.1 GEZM01091082 GEZM01091078 JAV57107.1 GDIQ01001129 JAN93608.1
Proteomes
UP000007151
UP000218220
UP000005204
UP000053268
UP000053240
UP000235965
+ More
UP000009046 UP000053825 UP000037069 UP000095301 UP000009192 UP000078542 UP000008237 UP000076502 UP000092443 UP000078200 UP000053097 UP000279307 UP000007755 UP000078540 UP000005205 UP000078492 UP000091820 UP000000311 UP000095300 UP000092445 UP000075809 UP000007819 UP000007798 UP000078541 UP000092460 UP000002358 UP000053105 UP000001292 UP000008711 UP000001070 UP000000803 UP000092553 UP000000304 UP000192223 UP000002282 UP000092444 UP000008820 UP000075883 UP000075900 UP000076408 UP000075885 UP000075920 UP000075884 UP000075886 UP000075880 UP000030765 UP000069272 UP000075903 UP000075840 UP000076407 UP000075882 UP000007062 UP000183832 UP000075902 UP000075881
UP000009046 UP000053825 UP000037069 UP000095301 UP000009192 UP000078542 UP000008237 UP000076502 UP000092443 UP000078200 UP000053097 UP000279307 UP000007755 UP000078540 UP000005205 UP000078492 UP000091820 UP000000311 UP000095300 UP000092445 UP000075809 UP000007819 UP000007798 UP000078541 UP000092460 UP000002358 UP000053105 UP000001292 UP000008711 UP000001070 UP000000803 UP000092553 UP000000304 UP000192223 UP000002282 UP000092444 UP000008820 UP000075883 UP000075900 UP000076408 UP000075885 UP000075920 UP000075884 UP000075886 UP000075880 UP000030765 UP000069272 UP000075903 UP000075840 UP000076407 UP000075882 UP000007062 UP000183832 UP000075902 UP000075881
Interpro
Gene 3D
ProteinModelPortal
O76340
A0A2H1VA46
A0A212F2T2
A0A2A4IYJ1
H9JMG0
A0A2A4IZH1
+ More
A0A194QCG4 A0A0N1PK87 A0A2J7PCG4 E0V9C1 A0A2J7PCG7 Q5FAN0 A0A0L7RKC4 A0A0L0BWC8 A0A1I8MPR9 A0A0Q9X803 A0A0Q9X9D2 A0A195CRM1 B4KDK3 E2BXW1 A0A154PK07 A0A1A9YAX6 A0A1A9VGU4 A0A026VSM1 F4X5Q2 A0A195BXI0 A0A158NUX0 A0A151IRN9 A0A1A9W1K2 E2B0Y5 A0A1I8P434 A0A1A9ZAA2 A0A151WSX9 J9K9E5 B4NKT4 A0A1I8P3Z3 A0A195FQ75 A0A1B0BM11 K7J2P7 A0A0M8ZVH0 B4HE17 B3P0Q9 B4JV23 Q8INF0 Q8INF0-2 A0A0M3QY21 B4QZ50 A0A1W4WIZ2 A0A1W4WIQ4 B4PS88 A0A1B0FDH5 Q16JH8 A0A182M001 A0A182RPV2 A0A182XVU3 A0A182PKI3 A0A182VYY9 A0A182NS81 A0A182QFQ0 A0A182JIS4 A0A084VHI9 A0A182F6T4 A0A182UMR7 A0A182I080 A0A182XBD8 A0A182KLU5 Q7PYK9 A0A1J1IFM2 A0A310SI97 A0A182TF11 A0A336LTP0 A0A1Y1K6M1 A0A1Y1KEA5 A0A182JZ34 A0A0P6IUP1
A0A194QCG4 A0A0N1PK87 A0A2J7PCG4 E0V9C1 A0A2J7PCG7 Q5FAN0 A0A0L7RKC4 A0A0L0BWC8 A0A1I8MPR9 A0A0Q9X803 A0A0Q9X9D2 A0A195CRM1 B4KDK3 E2BXW1 A0A154PK07 A0A1A9YAX6 A0A1A9VGU4 A0A026VSM1 F4X5Q2 A0A195BXI0 A0A158NUX0 A0A151IRN9 A0A1A9W1K2 E2B0Y5 A0A1I8P434 A0A1A9ZAA2 A0A151WSX9 J9K9E5 B4NKT4 A0A1I8P3Z3 A0A195FQ75 A0A1B0BM11 K7J2P7 A0A0M8ZVH0 B4HE17 B3P0Q9 B4JV23 Q8INF0 Q8INF0-2 A0A0M3QY21 B4QZ50 A0A1W4WIZ2 A0A1W4WIQ4 B4PS88 A0A1B0FDH5 Q16JH8 A0A182M001 A0A182RPV2 A0A182XVU3 A0A182PKI3 A0A182VYY9 A0A182NS81 A0A182QFQ0 A0A182JIS4 A0A084VHI9 A0A182F6T4 A0A182UMR7 A0A182I080 A0A182XBD8 A0A182KLU5 Q7PYK9 A0A1J1IFM2 A0A310SI97 A0A182TF11 A0A336LTP0 A0A1Y1K6M1 A0A1Y1KEA5 A0A182JZ34 A0A0P6IUP1
PDB
5O5L
E-value=9.23607e-44,
Score=449
Ontologies
KEGG
PATHWAY
GO
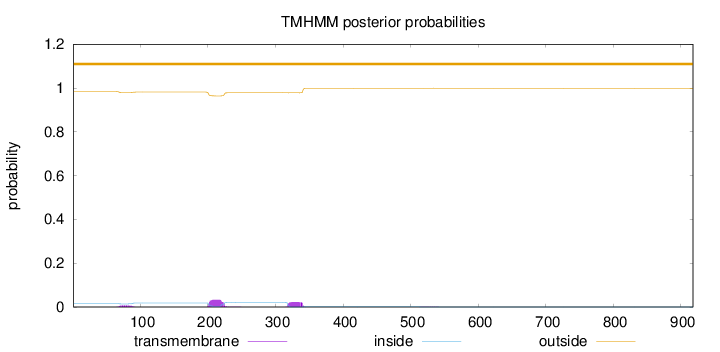
Topology
Subcellular location
Cytoplasm
Length:
918
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.34766
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.01564
outside
1 - 918
Population Genetic Test Statistics
Pi
206.677746
Theta
161.954715
Tajima's D
0.789221
CLR
0.268856
CSRT
0.597170141492925
Interpretation
Uncertain
