Gene
KWMTBOMO15622
Pre Gene Modal
BGIBMGA010842
Annotation
PREDICTED:_amine_sulfotransferase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.595
Sequence
CDS
ATGTTTGTTAAGAAGGATTGCATGGTCGAGATCAATCCTGGCCGGGTGATACTGCCGGCTGATTATATGACGATTGGATCGGATATATTGGATATGGAGGTACGGGAGAACGACGTTTGGATGTGCTCCTATCCGAGGACAGGTTCAACGTGGGCACAAGAGATGGTGTGGCTTATCGGGCACGACTTGGATTATGAAGGAGCCAAATCTTTACAGCAAATTCGATGCCCTTTAGTCGAGCTATCTTGTATTATGGTGGACGGGCATTCCGAATGGCATGACGAGTCAGTGAAGGGAACGTCAGTAGATTTGGTGAAGTACCGCCTTCCAGTACCTCGGTACATCCGGAGCCATCTTCCTTGGGACTTATTGCCGGTGGATATTCAGAGGGAAGACGGTTTTGCTAAGCCTAAGATTATATATACTTCTCGTAATCCGAAAGATATGGTTGTGTCGTATTACCACTACTGCACCCTAGTACACGGCCTGAAGGGTTCGTTTGAAGAGTTCTGCGACATCTTCATGAAGGATCATGCTCCTTTCGGACCAGTCTGGAACCATATTCTGGGTTTCTGGAATCGTCGTCACGATCCGAACATACTATTTATAAAGTTCGAAGAGATGAAACGTAACTTACCAGCTGTCGTGAGACGCACGGCGAAATTTCTCGAAAAGTCTCTTTCAGATGATGACGTCACCAAGCTATGCGATCACTTATCCTTCAGTAACATGAAGACTAACAGAGCTGTTAATCTTGAAGCTATATTGGAGAAATCATACGGGAAGAGCTACTTGGAAGCAACGGATTTACGATTCATAAGAAAAGGAGAAATTGGTGATTGGAAAAATTATATGTCCGATGATTTGTCTCGAAGTTTCGACGAATGGGCCGCGGAACATATAAAAGGCACCGGTCTGAGTTTTGAATAA
Protein
MFVKKDCMVEINPGRVILPADYMTIGSDILDMEVRENDVWMCSYPRTGSTWAQEMVWLIGHDLDYEGAKSLQQIRCPLVELSCIMVDGHSEWHDESVKGTSVDLVKYRLPVPRYIRSHLPWDLLPVDIQREDGFAKPKIIYTSRNPKDMVVSYYHYCTLVHGLKGSFEEFCDIFMKDHAPFGPVWNHILGFWNRRHDPNILFIKFEEMKRNLPAVVRRTAKFLEKSLSDDDVTKLCDHLSFSNMKTNRAVNLEAILEKSYGKSYLEATDLRFIRKGEIGDWKNYMSDDLSRSFDEWAAEHIKGTGLSFE
Summary
Uniprot
H9JMT9
A0A194QI78
A0A2H1VBA4
A0A212F2T0
A0A3S2LTW8
A0A0N0PE28
+ More
E0VVB6 A0A1B0Y0B0 A0A2J7PYP0 A0A0L7LRN9 A0A2P8ZP84 A0A1S4F7Z7 Q17CK0 A0A182GGR2 B0X1J9 A0A336L029 A0A336N8V6 A0A336MPF0 A0A0L0BR23 A0A034VUE6 A0A1A9VDQ3 A0A1B0G225 A0A1A9ZSW1 A0A1A9Y0I3 A0A0K8UI22 C4WUY7 A0A1I8NBP1 A0A0A1XGA6 B4NG18 R4FNY7 A0A1B6DEM3 A0A182RE41 A0A182PP76 B4JRK1 A0A023F9A7 A0A182V1Y9 A0A182LXK1 A0A182KB24 B4M0P6 A0A084WP52 A0A182HYP2 A0A182XII7 Q7QA04 B4KAN7 A0A182KZJ4 A0A0M4F5Z0 A0A182Y8S3 B3M1L8 A0A1I8P0S0 A0A182W3G1 B3P1S9 K7IVC8 A0A182F6M4 D6X2G0 B4PU10 A0A3B0K3G8 A0A1W4VRM0 B4QX90 A0A146MEA5 A0A1S3DFN2 Q9VHH0 A0A026WIM8 A0A146L0R4 B4HKH0 A0A151WW78 A0A182IP82 A0A158P0T9 Q29CF8 A0A195FYT4 A0A195AU22 A0A1B6MJW5 W5J6J3 A0A182R0P4 A0A182NGF7 F4WUV3 A0A195CZ25 E2AKF4 A0A161MN70 A0A0N0U4K4 A0A0L7QKU7 A0A088AF19 E2BE32 A0A1W4W294 A0A2A3EUM4 A0A154P9T0 A0A1B6KW79 A0A1B6IRV2 B4GN41 A0A1B0D7P0 A0A0L7K3K4 A0A232F7D1 A0A1J1J6P9 A0A1B6DTP8 A0A067RBR8 A0A2J7PYP3 E9IZB6 A0A2J7PYM4 A0A2J7PYQ0 A0A1B6EMJ4
E0VVB6 A0A1B0Y0B0 A0A2J7PYP0 A0A0L7LRN9 A0A2P8ZP84 A0A1S4F7Z7 Q17CK0 A0A182GGR2 B0X1J9 A0A336L029 A0A336N8V6 A0A336MPF0 A0A0L0BR23 A0A034VUE6 A0A1A9VDQ3 A0A1B0G225 A0A1A9ZSW1 A0A1A9Y0I3 A0A0K8UI22 C4WUY7 A0A1I8NBP1 A0A0A1XGA6 B4NG18 R4FNY7 A0A1B6DEM3 A0A182RE41 A0A182PP76 B4JRK1 A0A023F9A7 A0A182V1Y9 A0A182LXK1 A0A182KB24 B4M0P6 A0A084WP52 A0A182HYP2 A0A182XII7 Q7QA04 B4KAN7 A0A182KZJ4 A0A0M4F5Z0 A0A182Y8S3 B3M1L8 A0A1I8P0S0 A0A182W3G1 B3P1S9 K7IVC8 A0A182F6M4 D6X2G0 B4PU10 A0A3B0K3G8 A0A1W4VRM0 B4QX90 A0A146MEA5 A0A1S3DFN2 Q9VHH0 A0A026WIM8 A0A146L0R4 B4HKH0 A0A151WW78 A0A182IP82 A0A158P0T9 Q29CF8 A0A195FYT4 A0A195AU22 A0A1B6MJW5 W5J6J3 A0A182R0P4 A0A182NGF7 F4WUV3 A0A195CZ25 E2AKF4 A0A161MN70 A0A0N0U4K4 A0A0L7QKU7 A0A088AF19 E2BE32 A0A1W4W294 A0A2A3EUM4 A0A154P9T0 A0A1B6KW79 A0A1B6IRV2 B4GN41 A0A1B0D7P0 A0A0L7K3K4 A0A232F7D1 A0A1J1J6P9 A0A1B6DTP8 A0A067RBR8 A0A2J7PYP3 E9IZB6 A0A2J7PYM4 A0A2J7PYQ0 A0A1B6EMJ4
Pubmed
19121390
26354079
22118469
20566863
26227816
29403074
+ More
17510324 26483478 26108605 25348373 25315136 25830018 17994087 25474469 24438588 12364791 14747013 17210077 20966253 25244985 20075255 18362917 19820115 17550304 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 21347285 15632085 20920257 23761445 21719571 20798317 28648823 24845553 21282665
17510324 26483478 26108605 25348373 25315136 25830018 17994087 25474469 24438588 12364791 14747013 17210077 20966253 25244985 20075255 18362917 19820115 17550304 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 21347285 15632085 20920257 23761445 21719571 20798317 28648823 24845553 21282665
EMBL
BABH01016863
BABH01016864
BABH01016865
KQ459193
KPJ03156.1
ODYU01001452
+ More
SOQ37672.1 AGBW02010685 OWR48040.1 RSAL01000010 RVE53686.1 KQ459941 KPJ19145.1 DS235806 EEB17322.1 KT000402 ANI86000.1 NEVH01020342 PNF21458.1 JTDY01000233 KOB78140.1 PYGN01000005 PSN58317.1 CH477308 EAT44021.1 JXUM01062403 KQ562198 KXJ76430.1 DS232261 EDS38648.1 UFQS01001379 UFQT01001379 SSX10585.1 SSX30269.1 UFQT01003658 SSX35178.1 UFQT01001554 SSX30989.1 JRES01001482 KNC22530.1 GAKP01013804 JAC45148.1 CCAG010010758 GDHF01026000 GDHF01009156 JAI26314.1 JAI43158.1 ABLF02039910 AK341279 BAH71707.1 GBXI01004342 JAD09950.1 CH964251 EDW83235.1 ACPB03016231 GAHY01001109 JAA76401.1 GEDC01013171 JAS24127.1 CH916373 EDV94391.1 GBBI01001214 JAC17498.1 AXCM01001003 CH940650 EDW67338.1 ATLV01024906 KE525364 KFB51996.1 APCN01005395 AAAB01008898 EAA09115.3 CH933806 EDW16774.1 CP012526 ALC47387.1 CH902617 EDV43309.1 CH954181 EDV49817.1 AAZX01008576 KQ971372 EFA09867.1 CM000160 EDW96621.1 OUUW01000005 SPP80166.1 CM000364 EDX13661.1 GDHC01000860 JAQ17769.1 AE014297 BT125580 AAF54344.1 ADM26248.2 AHN57243.1 KK107199 QOIP01000009 EZA55511.1 RLU18559.1 GDHC01016455 JAQ02174.1 CH480815 EDW42920.1 KQ982691 KYQ52076.1 ADTU01005787 CM000070 EAL26688.2 KQ981200 KYN44989.1 KQ976738 KYM75681.1 GEBQ01017860 GEBQ01003751 JAT22117.1 JAT36226.1 ADMH02001987 ETN60077.1 AXCN02001608 GL888378 EGI62017.1 KQ977141 KYN05384.1 GL440262 EFN66096.1 GEMB01001328 JAS01828.1 KQ435821 KOX72374.1 KQ414940 KOC59248.1 GL447755 EFN86019.1 KZ288185 PBC34916.1 KQ434851 KZC08593.1 GEBQ01024261 JAT15716.1 GECU01018045 JAS89661.1 CH479186 EDW39167.1 AJVK01027063 AJVK01027064 JTDY01011848 KOB52376.1 NNAY01000789 OXU26502.1 CVRI01000070 CRL07158.1 GEDC01008255 JAS29043.1 KK852806 KDR16097.1 PNF21432.1 GL767121 EFZ14098.1 PNF21436.1 PNF21430.1 GECZ01030617 JAS39152.1
SOQ37672.1 AGBW02010685 OWR48040.1 RSAL01000010 RVE53686.1 KQ459941 KPJ19145.1 DS235806 EEB17322.1 KT000402 ANI86000.1 NEVH01020342 PNF21458.1 JTDY01000233 KOB78140.1 PYGN01000005 PSN58317.1 CH477308 EAT44021.1 JXUM01062403 KQ562198 KXJ76430.1 DS232261 EDS38648.1 UFQS01001379 UFQT01001379 SSX10585.1 SSX30269.1 UFQT01003658 SSX35178.1 UFQT01001554 SSX30989.1 JRES01001482 KNC22530.1 GAKP01013804 JAC45148.1 CCAG010010758 GDHF01026000 GDHF01009156 JAI26314.1 JAI43158.1 ABLF02039910 AK341279 BAH71707.1 GBXI01004342 JAD09950.1 CH964251 EDW83235.1 ACPB03016231 GAHY01001109 JAA76401.1 GEDC01013171 JAS24127.1 CH916373 EDV94391.1 GBBI01001214 JAC17498.1 AXCM01001003 CH940650 EDW67338.1 ATLV01024906 KE525364 KFB51996.1 APCN01005395 AAAB01008898 EAA09115.3 CH933806 EDW16774.1 CP012526 ALC47387.1 CH902617 EDV43309.1 CH954181 EDV49817.1 AAZX01008576 KQ971372 EFA09867.1 CM000160 EDW96621.1 OUUW01000005 SPP80166.1 CM000364 EDX13661.1 GDHC01000860 JAQ17769.1 AE014297 BT125580 AAF54344.1 ADM26248.2 AHN57243.1 KK107199 QOIP01000009 EZA55511.1 RLU18559.1 GDHC01016455 JAQ02174.1 CH480815 EDW42920.1 KQ982691 KYQ52076.1 ADTU01005787 CM000070 EAL26688.2 KQ981200 KYN44989.1 KQ976738 KYM75681.1 GEBQ01017860 GEBQ01003751 JAT22117.1 JAT36226.1 ADMH02001987 ETN60077.1 AXCN02001608 GL888378 EGI62017.1 KQ977141 KYN05384.1 GL440262 EFN66096.1 GEMB01001328 JAS01828.1 KQ435821 KOX72374.1 KQ414940 KOC59248.1 GL447755 EFN86019.1 KZ288185 PBC34916.1 KQ434851 KZC08593.1 GEBQ01024261 JAT15716.1 GECU01018045 JAS89661.1 CH479186 EDW39167.1 AJVK01027063 AJVK01027064 JTDY01011848 KOB52376.1 NNAY01000789 OXU26502.1 CVRI01000070 CRL07158.1 GEDC01008255 JAS29043.1 KK852806 KDR16097.1 PNF21432.1 GL767121 EFZ14098.1 PNF21436.1 PNF21430.1 GECZ01030617 JAS39152.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000283053
UP000053240
UP000009046
+ More
UP000235965 UP000037510 UP000245037 UP000008820 UP000069940 UP000249989 UP000002320 UP000037069 UP000078200 UP000092444 UP000092445 UP000092443 UP000007819 UP000095301 UP000007798 UP000015103 UP000075900 UP000075885 UP000001070 UP000075903 UP000075883 UP000075881 UP000008792 UP000030765 UP000075840 UP000076407 UP000007062 UP000009192 UP000075882 UP000092553 UP000076408 UP000007801 UP000095300 UP000075920 UP000008711 UP000002358 UP000069272 UP000007266 UP000002282 UP000268350 UP000192221 UP000000304 UP000079169 UP000000803 UP000053097 UP000279307 UP000001292 UP000075809 UP000075880 UP000005205 UP000001819 UP000078541 UP000078540 UP000000673 UP000075886 UP000075884 UP000007755 UP000078542 UP000000311 UP000053105 UP000053825 UP000005203 UP000008237 UP000242457 UP000076502 UP000008744 UP000092462 UP000215335 UP000183832 UP000027135
UP000235965 UP000037510 UP000245037 UP000008820 UP000069940 UP000249989 UP000002320 UP000037069 UP000078200 UP000092444 UP000092445 UP000092443 UP000007819 UP000095301 UP000007798 UP000015103 UP000075900 UP000075885 UP000001070 UP000075903 UP000075883 UP000075881 UP000008792 UP000030765 UP000075840 UP000076407 UP000007062 UP000009192 UP000075882 UP000092553 UP000076408 UP000007801 UP000095300 UP000075920 UP000008711 UP000002358 UP000069272 UP000007266 UP000002282 UP000268350 UP000192221 UP000000304 UP000079169 UP000000803 UP000053097 UP000279307 UP000001292 UP000075809 UP000075880 UP000005205 UP000001819 UP000078541 UP000078540 UP000000673 UP000075886 UP000075884 UP000007755 UP000078542 UP000000311 UP000053105 UP000053825 UP000005203 UP000008237 UP000242457 UP000076502 UP000008744 UP000092462 UP000215335 UP000183832 UP000027135
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JMT9
A0A194QI78
A0A2H1VBA4
A0A212F2T0
A0A3S2LTW8
A0A0N0PE28
+ More
E0VVB6 A0A1B0Y0B0 A0A2J7PYP0 A0A0L7LRN9 A0A2P8ZP84 A0A1S4F7Z7 Q17CK0 A0A182GGR2 B0X1J9 A0A336L029 A0A336N8V6 A0A336MPF0 A0A0L0BR23 A0A034VUE6 A0A1A9VDQ3 A0A1B0G225 A0A1A9ZSW1 A0A1A9Y0I3 A0A0K8UI22 C4WUY7 A0A1I8NBP1 A0A0A1XGA6 B4NG18 R4FNY7 A0A1B6DEM3 A0A182RE41 A0A182PP76 B4JRK1 A0A023F9A7 A0A182V1Y9 A0A182LXK1 A0A182KB24 B4M0P6 A0A084WP52 A0A182HYP2 A0A182XII7 Q7QA04 B4KAN7 A0A182KZJ4 A0A0M4F5Z0 A0A182Y8S3 B3M1L8 A0A1I8P0S0 A0A182W3G1 B3P1S9 K7IVC8 A0A182F6M4 D6X2G0 B4PU10 A0A3B0K3G8 A0A1W4VRM0 B4QX90 A0A146MEA5 A0A1S3DFN2 Q9VHH0 A0A026WIM8 A0A146L0R4 B4HKH0 A0A151WW78 A0A182IP82 A0A158P0T9 Q29CF8 A0A195FYT4 A0A195AU22 A0A1B6MJW5 W5J6J3 A0A182R0P4 A0A182NGF7 F4WUV3 A0A195CZ25 E2AKF4 A0A161MN70 A0A0N0U4K4 A0A0L7QKU7 A0A088AF19 E2BE32 A0A1W4W294 A0A2A3EUM4 A0A154P9T0 A0A1B6KW79 A0A1B6IRV2 B4GN41 A0A1B0D7P0 A0A0L7K3K4 A0A232F7D1 A0A1J1J6P9 A0A1B6DTP8 A0A067RBR8 A0A2J7PYP3 E9IZB6 A0A2J7PYM4 A0A2J7PYQ0 A0A1B6EMJ4
E0VVB6 A0A1B0Y0B0 A0A2J7PYP0 A0A0L7LRN9 A0A2P8ZP84 A0A1S4F7Z7 Q17CK0 A0A182GGR2 B0X1J9 A0A336L029 A0A336N8V6 A0A336MPF0 A0A0L0BR23 A0A034VUE6 A0A1A9VDQ3 A0A1B0G225 A0A1A9ZSW1 A0A1A9Y0I3 A0A0K8UI22 C4WUY7 A0A1I8NBP1 A0A0A1XGA6 B4NG18 R4FNY7 A0A1B6DEM3 A0A182RE41 A0A182PP76 B4JRK1 A0A023F9A7 A0A182V1Y9 A0A182LXK1 A0A182KB24 B4M0P6 A0A084WP52 A0A182HYP2 A0A182XII7 Q7QA04 B4KAN7 A0A182KZJ4 A0A0M4F5Z0 A0A182Y8S3 B3M1L8 A0A1I8P0S0 A0A182W3G1 B3P1S9 K7IVC8 A0A182F6M4 D6X2G0 B4PU10 A0A3B0K3G8 A0A1W4VRM0 B4QX90 A0A146MEA5 A0A1S3DFN2 Q9VHH0 A0A026WIM8 A0A146L0R4 B4HKH0 A0A151WW78 A0A182IP82 A0A158P0T9 Q29CF8 A0A195FYT4 A0A195AU22 A0A1B6MJW5 W5J6J3 A0A182R0P4 A0A182NGF7 F4WUV3 A0A195CZ25 E2AKF4 A0A161MN70 A0A0N0U4K4 A0A0L7QKU7 A0A088AF19 E2BE32 A0A1W4W294 A0A2A3EUM4 A0A154P9T0 A0A1B6KW79 A0A1B6IRV2 B4GN41 A0A1B0D7P0 A0A0L7K3K4 A0A232F7D1 A0A1J1J6P9 A0A1B6DTP8 A0A067RBR8 A0A2J7PYP3 E9IZB6 A0A2J7PYM4 A0A2J7PYQ0 A0A1B6EMJ4
PDB
1FML
E-value=1.27967e-43,
Score=443
Ontologies
GO
Topology
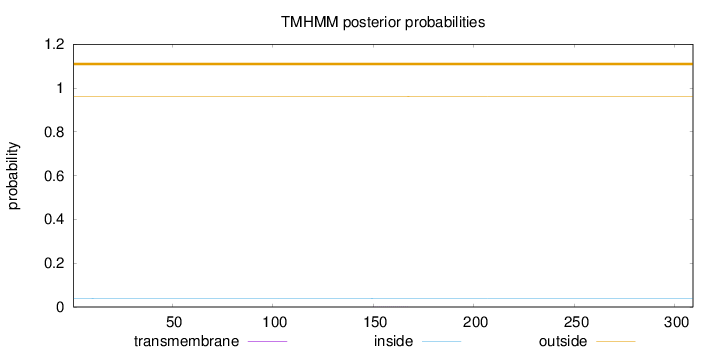
Length:
309
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00657
Exp number, first 60 AAs:
0.00211
Total prob of N-in:
0.03964
outside
1 - 309
Population Genetic Test Statistics
Pi
152.719805
Theta
166.293049
Tajima's D
0.28916
CLR
10.992912
CSRT
0.451627418629069
Interpretation
Uncertain
