Gene
KWMTBOMO15618
Pre Gene Modal
BGIBMGA010715
Annotation
PREDICTED:_dynein_light_chain_roadblock-type_2_[Amyelois_transitella]
Full name
Dynein light chain roadblock
Location in the cell
Cytoplasmic Reliability : 1.695
Sequence
CDS
ATGGCCAATGAAGCTGAGGACACAATTCGAAAACTGTCGACGTACAAAGGCGTGGTTGGCGTCGTCGTTGTAAACGGAGAAGGAATACCGATAAAAACCACTTTAGATAATCACATTTCCGTACAATACGCGGGCCTATTGACCGCGTTGGTGGACAAAGCGAAAACTGTCGTGCGAGACCTGGATCCGTCTAATGACTTGACATTTTTGAGGATAAGAAGCAAAAAAAGCGAAGTAATGGTGGCGCCCGATAAAGAATTTATATTAATAGTAGTTCAAAATCCTGTTGAGTAA
Protein
MANEAEDTIRKLSTYKGVVGVVVVNGEGIPIKTTLDNHISVQYAGLLTALVDKAKTVVRDLDPSNDLTFLRIRSKKSEVMVAPDKEFILIVVQNPVE
Summary
Description
Acts as one of several non-catalytic accessory components of the cytoplasmic dynein 1 complex that are thought to be involved in linking dynein to cargos and to adapter proteins that regulate dynein function. Cytoplasmic dynein 1 acts as a motor for the intracellular retrograde motility of vesicles and organelles along microtubules.
Subunit
Homodimer. The cytoplasmic dynein 1 complex consists of two catalytic heavy chains (HCs) and a number of non-catalytic subunits presented by intermediate chains (ICs), light intermediate chains (LICs) and light chains (LCs); the composition seems to vary in respect to the IC, LIC and LC composition. The heavy chain homodimer serves as a scaffold for the probable homodimeric assembly of the respective non-catalytic subunits. The ICs and LICs bind directly to the HC dimer and the LCs assemble on the IC dimer.
Similarity
Belongs to the GAMAD family.
Uniprot
H9JMG2
A0A194QDX9
A0A0N1IC30
A0A2A4IYT4
A0A212F2U0
A0A2H1W2P3
+ More
A0A0L7LRZ1 J3JTY4 A0A1W4X676 D6WQQ5 K7JA95 A0A0J7KY16 A0A026WJ25 A0A2A3EMM9 A0A088ADJ6 A0A067RH23 A0A1B6KJ82 A0A158NYD5 A0A1B6F9X3 A0A195AYE1 A0A2S2PVG4 A0A151WJ12 A0A151JRJ4 E2BRK4 A0A151IN50 A0A154PC50 A0A2S2PD67 A0A2H8TPR3 C4WVV2 E0W0E9 A0A1L8DX00 A0A1W4VCE7 B4LPW2 B4KQZ2 A0A3B0J522 Q291G9 B4GAR3 B4P7Z5 A0A0M5IXC9 B3MG72 B4NNF3 B4J925 B3NNR1 B4QAU0 B4HMM0 Q7KMS3 A0A0K8TPR0 A0A1B0GJT0 A0A1I8PM73 A0A1L8EGK4 A0A1I8MN78 T1D5K7 A0A1Q3FIA4 B0W4E5 Q17AA0 A0A023ECK0 U5EKI8 A0A1Y2HZV2 A0A3B0JPV3 T1DQJ3 A0A2M3ZIQ8 A0A2M4AQW6 A0A182MI57 A0A182PCW5 A0A182V2N0 W5JFP9 A0A182K785 A0A182VYA3 A0A182XKC0 Q5TSL8 A0A182HJM2 D3TRW5 A0A182QHH1 A0A1Y2ECB2 A0A224Y1Q6 A0A0V0G8H2 A0A069DNV8 I3MWU1 A0A2K6KE40 A0A2K6PVH6 A0A1Y2ATV3 A0A3P9AMS4 A0A0L0RWQ7 A0A023FAQ0 A0A182XWL5 A0A182RFX8 M3W1J3 A0A139AX07 A0A0P4VTR1 R4FMK9 F7B1T3 W5LEA2 A0A0L7RGT0 F4WSY8 A0A195F756 E2AZ45 A0A1S3KPY1 C1BGV4 D4A0A0 E2RQD9
A0A0L7LRZ1 J3JTY4 A0A1W4X676 D6WQQ5 K7JA95 A0A0J7KY16 A0A026WJ25 A0A2A3EMM9 A0A088ADJ6 A0A067RH23 A0A1B6KJ82 A0A158NYD5 A0A1B6F9X3 A0A195AYE1 A0A2S2PVG4 A0A151WJ12 A0A151JRJ4 E2BRK4 A0A151IN50 A0A154PC50 A0A2S2PD67 A0A2H8TPR3 C4WVV2 E0W0E9 A0A1L8DX00 A0A1W4VCE7 B4LPW2 B4KQZ2 A0A3B0J522 Q291G9 B4GAR3 B4P7Z5 A0A0M5IXC9 B3MG72 B4NNF3 B4J925 B3NNR1 B4QAU0 B4HMM0 Q7KMS3 A0A0K8TPR0 A0A1B0GJT0 A0A1I8PM73 A0A1L8EGK4 A0A1I8MN78 T1D5K7 A0A1Q3FIA4 B0W4E5 Q17AA0 A0A023ECK0 U5EKI8 A0A1Y2HZV2 A0A3B0JPV3 T1DQJ3 A0A2M3ZIQ8 A0A2M4AQW6 A0A182MI57 A0A182PCW5 A0A182V2N0 W5JFP9 A0A182K785 A0A182VYA3 A0A182XKC0 Q5TSL8 A0A182HJM2 D3TRW5 A0A182QHH1 A0A1Y2ECB2 A0A224Y1Q6 A0A0V0G8H2 A0A069DNV8 I3MWU1 A0A2K6KE40 A0A2K6PVH6 A0A1Y2ATV3 A0A3P9AMS4 A0A0L0RWQ7 A0A023FAQ0 A0A182XWL5 A0A182RFX8 M3W1J3 A0A139AX07 A0A0P4VTR1 R4FMK9 F7B1T3 W5LEA2 A0A0L7RGT0 F4WSY8 A0A195F756 E2AZ45 A0A1S3KPY1 C1BGV4 D4A0A0 E2RQD9
Pubmed
19121390
26354079
22118469
26227816
22516182
23537049
+ More
18362917 19820115 20075255 24508170 30249741 24845553 21347285 20798317 20566863 17994087 15632085 17550304 22936249 10402468 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 25315136 24330624 17510324 24945155 20920257 23761445 12364791 14747013 17210077 20353571 26334808 25362486 25069045 25474469 25244985 17975172 25977457 27129103 19892987 25329095 21719571 15057822 15632090 16341006
18362917 19820115 20075255 24508170 30249741 24845553 21347285 20798317 20566863 17994087 15632085 17550304 22936249 10402468 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 25315136 24330624 17510324 24945155 20920257 23761445 12364791 14747013 17210077 20353571 26334808 25362486 25069045 25474469 25244985 17975172 25977457 27129103 19892987 25329095 21719571 15057822 15632090 16341006
EMBL
BABH01016863
KQ459193
KPJ03155.1
KQ459941
KPJ19144.1
NWSH01004564
+ More
PCG64921.1 AGBW02010685 OWR48046.1 ODYU01005946 SOQ47365.1 JTDY01000233 KOB78144.1 APGK01054226 BT126694 KB741248 KB632305 AEE61656.1 ENN71942.1 ERL91859.1 KQ971354 EFA07590.1 AAZX01013217 AAZX01014215 LBMM01001988 KMQ95442.1 KK107178 QOIP01000006 EZA56047.1 RLU21236.1 KZ288206 PBC33063.1 KK852686 KDR18463.1 GEBQ01028481 JAT11496.1 ADTU01003759 GECZ01022743 JAS47026.1 KQ976701 KYM77216.1 GGMS01000325 MBY69528.1 KQ983055 KYQ47814.1 KQ978607 KYN29914.1 GL449965 EFN81696.1 KQ976963 KYN06777.1 KQ434864 KZC09004.1 GGMR01014784 MBY27403.1 GFXV01000619 GFXV01003827 MBW12424.1 MBW15632.1 ABLF02022556 AK341692 BAH72022.1 DS235858 EEB19105.1 GFDF01003101 JAV10983.1 CH940648 EDW61302.1 CH933808 EDW10348.2 OUUW01000001 SPP74682.1 CM000071 EAL25143.1 CH479181 EDW32015.1 CM000158 EDW92150.1 CP012524 ALC41081.1 CH902619 EDV36767.1 CH964282 EDW85892.1 CH916367 EDW01374.1 CH954179 EDV55618.2 CM000362 CM002911 EDX07491.1 KMY94544.1 CH480816 CH481394 EDW48288.1 EDW54727.1 AF141920 AF141921 AE013599 AY071587 KX532053 AAD45987.1 AAF57861.1 AAL49209.1 ANY27863.1 GDAI01001485 JAI16118.1 AJWK01022211 GFDG01000949 JAV17850.1 GALA01000463 JAA94389.1 GFDL01007812 JAV27233.1 DS231836 EDS33230.1 CH477337 EAT43191.1 GAPW01006516 GAPW01006515 GEHC01000027 JAC07082.1 JAV47618.1 GANO01001797 JAB58074.1 MCFL01000003 ORZ40135.1 SPP74681.1 GAMD01002449 JAA99141.1 GGFM01007638 MBW28389.1 GGFK01009842 MBW43163.1 AXCM01006045 ADMH02001529 GGFL01004252 ETN62168.1 MBW68430.1 AAAB01008904 EAA09669.4 APCN01002188 EZ424167 ADD20443.1 AXCN02000300 MCOG01000045 ORY69208.1 GFTR01001474 JAW14952.1 GECL01001689 JAP04435.1 GBGD01003612 JAC85277.1 AGTP01010344 MCOG01000206 ORY26003.1 GG745328 GG745330 KNE54539.1 KNE56187.1 GBBI01000729 GEMB01001187 JAC17983.1 JAS01961.1 AANG04002512 KQ965733 KXS21239.1 GDKW01000570 JAI56025.1 ACPB03005914 GAHY01000948 JAA76562.1 KQ414596 KOC70038.1 GL888331 EGI62692.1 KQ981768 KYN36017.1 GL444080 EFN61303.1 BT073833 BT074039 BT074166 ACO08257.1 AABR07043941 AABR07043942 CH473972 EDL92633.1 AAEX03004043
PCG64921.1 AGBW02010685 OWR48046.1 ODYU01005946 SOQ47365.1 JTDY01000233 KOB78144.1 APGK01054226 BT126694 KB741248 KB632305 AEE61656.1 ENN71942.1 ERL91859.1 KQ971354 EFA07590.1 AAZX01013217 AAZX01014215 LBMM01001988 KMQ95442.1 KK107178 QOIP01000006 EZA56047.1 RLU21236.1 KZ288206 PBC33063.1 KK852686 KDR18463.1 GEBQ01028481 JAT11496.1 ADTU01003759 GECZ01022743 JAS47026.1 KQ976701 KYM77216.1 GGMS01000325 MBY69528.1 KQ983055 KYQ47814.1 KQ978607 KYN29914.1 GL449965 EFN81696.1 KQ976963 KYN06777.1 KQ434864 KZC09004.1 GGMR01014784 MBY27403.1 GFXV01000619 GFXV01003827 MBW12424.1 MBW15632.1 ABLF02022556 AK341692 BAH72022.1 DS235858 EEB19105.1 GFDF01003101 JAV10983.1 CH940648 EDW61302.1 CH933808 EDW10348.2 OUUW01000001 SPP74682.1 CM000071 EAL25143.1 CH479181 EDW32015.1 CM000158 EDW92150.1 CP012524 ALC41081.1 CH902619 EDV36767.1 CH964282 EDW85892.1 CH916367 EDW01374.1 CH954179 EDV55618.2 CM000362 CM002911 EDX07491.1 KMY94544.1 CH480816 CH481394 EDW48288.1 EDW54727.1 AF141920 AF141921 AE013599 AY071587 KX532053 AAD45987.1 AAF57861.1 AAL49209.1 ANY27863.1 GDAI01001485 JAI16118.1 AJWK01022211 GFDG01000949 JAV17850.1 GALA01000463 JAA94389.1 GFDL01007812 JAV27233.1 DS231836 EDS33230.1 CH477337 EAT43191.1 GAPW01006516 GAPW01006515 GEHC01000027 JAC07082.1 JAV47618.1 GANO01001797 JAB58074.1 MCFL01000003 ORZ40135.1 SPP74681.1 GAMD01002449 JAA99141.1 GGFM01007638 MBW28389.1 GGFK01009842 MBW43163.1 AXCM01006045 ADMH02001529 GGFL01004252 ETN62168.1 MBW68430.1 AAAB01008904 EAA09669.4 APCN01002188 EZ424167 ADD20443.1 AXCN02000300 MCOG01000045 ORY69208.1 GFTR01001474 JAW14952.1 GECL01001689 JAP04435.1 GBGD01003612 JAC85277.1 AGTP01010344 MCOG01000206 ORY26003.1 GG745328 GG745330 KNE54539.1 KNE56187.1 GBBI01000729 GEMB01001187 JAC17983.1 JAS01961.1 AANG04002512 KQ965733 KXS21239.1 GDKW01000570 JAI56025.1 ACPB03005914 GAHY01000948 JAA76562.1 KQ414596 KOC70038.1 GL888331 EGI62692.1 KQ981768 KYN36017.1 GL444080 EFN61303.1 BT073833 BT074039 BT074166 ACO08257.1 AABR07043941 AABR07043942 CH473972 EDL92633.1 AAEX03004043
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000019118 UP000030742 UP000192223 UP000007266 UP000002358 UP000036403 UP000053097 UP000279307 UP000242457 UP000005203 UP000027135 UP000005205 UP000078540 UP000075809 UP000078492 UP000008237 UP000078542 UP000076502 UP000007819 UP000009046 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000008744 UP000002282 UP000092553 UP000007801 UP000007798 UP000001070 UP000008711 UP000000304 UP000001292 UP000000803 UP000092461 UP000095300 UP000095301 UP000002320 UP000008820 UP000193411 UP000075883 UP000075885 UP000075903 UP000000673 UP000075881 UP000075920 UP000076407 UP000007062 UP000075840 UP000075886 UP000193920 UP000005215 UP000233180 UP000233200 UP000265140 UP000054350 UP000076408 UP000075900 UP000011712 UP000070544 UP000015103 UP000002281 UP000018467 UP000053825 UP000007755 UP000078541 UP000000311 UP000087266 UP000002494 UP000002254
UP000019118 UP000030742 UP000192223 UP000007266 UP000002358 UP000036403 UP000053097 UP000279307 UP000242457 UP000005203 UP000027135 UP000005205 UP000078540 UP000075809 UP000078492 UP000008237 UP000078542 UP000076502 UP000007819 UP000009046 UP000192221 UP000008792 UP000009192 UP000268350 UP000001819 UP000008744 UP000002282 UP000092553 UP000007801 UP000007798 UP000001070 UP000008711 UP000000304 UP000001292 UP000000803 UP000092461 UP000095300 UP000095301 UP000002320 UP000008820 UP000193411 UP000075883 UP000075885 UP000075903 UP000000673 UP000075881 UP000075920 UP000076407 UP000007062 UP000075840 UP000075886 UP000193920 UP000005215 UP000233180 UP000233200 UP000265140 UP000054350 UP000076408 UP000075900 UP000011712 UP000070544 UP000015103 UP000002281 UP000018467 UP000053825 UP000007755 UP000078541 UP000000311 UP000087266 UP000002494 UP000002254
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JMG2
A0A194QDX9
A0A0N1IC30
A0A2A4IYT4
A0A212F2U0
A0A2H1W2P3
+ More
A0A0L7LRZ1 J3JTY4 A0A1W4X676 D6WQQ5 K7JA95 A0A0J7KY16 A0A026WJ25 A0A2A3EMM9 A0A088ADJ6 A0A067RH23 A0A1B6KJ82 A0A158NYD5 A0A1B6F9X3 A0A195AYE1 A0A2S2PVG4 A0A151WJ12 A0A151JRJ4 E2BRK4 A0A151IN50 A0A154PC50 A0A2S2PD67 A0A2H8TPR3 C4WVV2 E0W0E9 A0A1L8DX00 A0A1W4VCE7 B4LPW2 B4KQZ2 A0A3B0J522 Q291G9 B4GAR3 B4P7Z5 A0A0M5IXC9 B3MG72 B4NNF3 B4J925 B3NNR1 B4QAU0 B4HMM0 Q7KMS3 A0A0K8TPR0 A0A1B0GJT0 A0A1I8PM73 A0A1L8EGK4 A0A1I8MN78 T1D5K7 A0A1Q3FIA4 B0W4E5 Q17AA0 A0A023ECK0 U5EKI8 A0A1Y2HZV2 A0A3B0JPV3 T1DQJ3 A0A2M3ZIQ8 A0A2M4AQW6 A0A182MI57 A0A182PCW5 A0A182V2N0 W5JFP9 A0A182K785 A0A182VYA3 A0A182XKC0 Q5TSL8 A0A182HJM2 D3TRW5 A0A182QHH1 A0A1Y2ECB2 A0A224Y1Q6 A0A0V0G8H2 A0A069DNV8 I3MWU1 A0A2K6KE40 A0A2K6PVH6 A0A1Y2ATV3 A0A3P9AMS4 A0A0L0RWQ7 A0A023FAQ0 A0A182XWL5 A0A182RFX8 M3W1J3 A0A139AX07 A0A0P4VTR1 R4FMK9 F7B1T3 W5LEA2 A0A0L7RGT0 F4WSY8 A0A195F756 E2AZ45 A0A1S3KPY1 C1BGV4 D4A0A0 E2RQD9
A0A0L7LRZ1 J3JTY4 A0A1W4X676 D6WQQ5 K7JA95 A0A0J7KY16 A0A026WJ25 A0A2A3EMM9 A0A088ADJ6 A0A067RH23 A0A1B6KJ82 A0A158NYD5 A0A1B6F9X3 A0A195AYE1 A0A2S2PVG4 A0A151WJ12 A0A151JRJ4 E2BRK4 A0A151IN50 A0A154PC50 A0A2S2PD67 A0A2H8TPR3 C4WVV2 E0W0E9 A0A1L8DX00 A0A1W4VCE7 B4LPW2 B4KQZ2 A0A3B0J522 Q291G9 B4GAR3 B4P7Z5 A0A0M5IXC9 B3MG72 B4NNF3 B4J925 B3NNR1 B4QAU0 B4HMM0 Q7KMS3 A0A0K8TPR0 A0A1B0GJT0 A0A1I8PM73 A0A1L8EGK4 A0A1I8MN78 T1D5K7 A0A1Q3FIA4 B0W4E5 Q17AA0 A0A023ECK0 U5EKI8 A0A1Y2HZV2 A0A3B0JPV3 T1DQJ3 A0A2M3ZIQ8 A0A2M4AQW6 A0A182MI57 A0A182PCW5 A0A182V2N0 W5JFP9 A0A182K785 A0A182VYA3 A0A182XKC0 Q5TSL8 A0A182HJM2 D3TRW5 A0A182QHH1 A0A1Y2ECB2 A0A224Y1Q6 A0A0V0G8H2 A0A069DNV8 I3MWU1 A0A2K6KE40 A0A2K6PVH6 A0A1Y2ATV3 A0A3P9AMS4 A0A0L0RWQ7 A0A023FAQ0 A0A182XWL5 A0A182RFX8 M3W1J3 A0A139AX07 A0A0P4VTR1 R4FMK9 F7B1T3 W5LEA2 A0A0L7RGT0 F4WSY8 A0A195F756 E2AZ45 A0A1S3KPY1 C1BGV4 D4A0A0 E2RQD9
PDB
3L9K
E-value=1.76042e-24,
Score=272
Ontologies
GO
Topology
Subcellular location
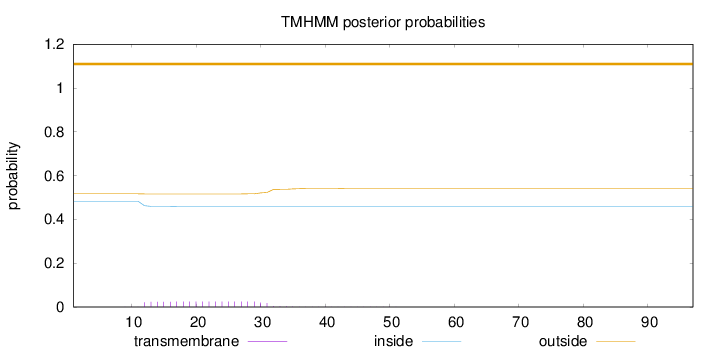
Length:
97
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.48911
Exp number, first 60 AAs:
0.48874
Total prob of N-in:
0.48330
outside
1 - 97
Population Genetic Test Statistics
Pi
217.086736
Theta
188.284286
Tajima's D
0.48139
CLR
6.914161
CSRT
0.503424828758562
Interpretation
Uncertain
