Gene
KWMTBOMO15617 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010840
Annotation
MLE_protein_[Bombyx_mori]
Full name
Dosage compensation regulator
Alternative Name
ATP-dependent RNA helicase mle
Protein male-less
Protein maleless
Protein no action potential
Protein male-less
Protein maleless
Protein no action potential
Location in the cell
Nuclear Reliability : 2.257
Sequence
CDS
ATGGATATTAAATCATTTTTATATACTTGGTGTGCTAAGAAAGGCTTAACTCCGCTTTTCGATATCCGGGCCACAGGTCCTAAACACCGCCAACGCTTTCTGTGCGAAGTCCGCGTCGAAGGATTTTCATATTTAGGAGCTGGAAACTCTACTACCAAAAAAGATGCACAGATGAACGCCGCTAAGGACTTTGTTAGTTATCTAGTTAGAGCGGGGCAAATCCCACAGGCTGATGTTCCTGAAGATGTGAAGGCAAAGGCAATTGAGGAATCTACTAATGAAGAATCTGTCGAGAATAATGGTGGTCAACAACCTAAGCAAGTCTTTCAGGATGGAATGGGTCCAGAAGTTATAGGACCAGCGTACCAAGCTTATGGAGGTGAAAATGACAAGAAATTTTCATACATAGATCGCATGCAACAACAGAAAAATGTTGAAGAAGCTGAAGATCTGGACATAAACTCCAGTCTCCATGGCAACTGGACAATGGAGAATGCTAAGTCAAAACTTCATCAGTTTTTACAGATGAATAAAATAAATGCAGACTATGTTTACAAAGCAGTTGGACCTGATCATACACGGAGTTTCGCTTGTGAGCTAACAATATATGTGAGTCAATTGGGACGCAATGTCACCGGTCGGGAGACCGCCTCAAACAAGCAAACCGCCAGCAAGTCATGTGCACTGTCGATCGTGAGACAGCTATACCATCTTGGAGTCATTGAGGCATACAGCGGTACGCTAAAAAGGGACAGATCCTCAGAGGGTATTATGAAACCATATCCAGTTGCTATATGCCCGGAACTTGAACAACAATTAGAAGATACCTTACAAGAATTAGAAATCGAGCCAGTTAATGTCGACGCACCAAGCCAAGATAACCAAGAGGGGAGTGCTGTGACTCTGTTGCAAAAAGATCGCGCGAAGGCTATGAGAGAAGCGAAGCCTATGCCGGCTGGTGTTGTGACGTGGTCTCCTCCGCAGCCGAATTGGAACGCTTGGACAGGGTGTAACATCGACGAGGGCGCTTTAGCAACTGCCTCCATGGACGAGATTAGCGCTGACCTGGAGAAATGTCACAGGAATATTTGTCAGAATAGCAAATTGTACCAGGAAAGCCTCAATGAAAGAGAACATTTGCCGGTGTTCTCCATGAAATCACAGATAATGGAAGCGATCAACGAGAACCCTGTGATCATCATCCGTGGTAATACTGGCTGCGGTAAAACTACACAGGTGTGCCAGTTCATCCTCGACGACTACATAGCGAGCGGTCAGGGCGCGTGGGCCAACGTGTGCGTGACGCAACCGCGCCGAATCTCCGCCGTCAGTGTCGCCGAGCGCGTCGCGGCTGAGAGGTGCGAAGAGCTCGGGAACACCGTCGGGTATTCAGTGCGTTTCGAGTCAGTGCTACCGCGGCCTTATGGGTCAATCATGTTCTGTACCGTCGGGGTGCTGCTGAGGAAACTGGAAGGCGGTCTGAGGGGTGTCAGCCACGTTTTAGTCGACGAGGTACACGAACGCGATGCCGATACGGACTTTGCTCTGATCCTGCTGCGAGACATGGCCCACACCTACCCCGACCTGCGCATCATTCTGATGTCGGCCACGGTCGACACCACGCTATTCGTAAAGTACTTCGGAGGCTGTCCAGTCATTGAGGTGCCAGGTCGAACGTATCCAGTGACGCAGTACTTCTTGGAAGACTCGATAGAGCTGACCAAGTTCATGCCCCCGCCGATCACTAGGAAACGGAAATCTACAGGAAAACGAGCGAATAAAGACGACGAAGATGACGACGACGAGGATGATTTGGATGAGCCTTACGAAGATTTGAACAAGCAATGCAGCTTGGGAGACGGATACTCTCAAGCCACAGTGGACGCTTTACAACAGTTGTCCGAGAGAGATTTCAGCTTCGAGTTGGTCCAAGCGATCCTCATGTACATCGATGGACTGGGAGGCGACGGCGCTGTCCTCGTGTTCCTCCCCGGGTGGAACCTCATCTTCGCGCTGATGAAGCACCTGTTGCAGCACAGACTCTTTGGAGATCCATCAAAATACGTTATACTGCCTCTGCACAGTCAAATACCGAGGGAGGATCAAAAGAAAGTGTTCATTACCCCACCGGAAGGGATCACTAAAGTGATCCTGTCAACCAACATAGCGGAGACTTCGATAACCATCAACGACGTGGTATACGTCATCGACTCGTGCAAGGCGAAAATGAAGCTGTTCACGTCCCACAACAACATGACGTCATACGCCACCGTATGGGCGAGTAAGACCAACCTCGAACAGCGCAAGGGGCGTGCGGGGCGCGTGCGGCCCGGCGTGTGCTTCACGCTGTGCACGTACGCGCGCTACGAGAAACTTGAGGAACATTTGGCCGCCGAAATGTTTAGGACTCCGCTACACGAGCTTGCTTTGAGCATAAAGCTCCTCCGCCTGGGTGCGATCGGTCACTTCCTGTCGAAGGCCCCGGAGCCCCCGCCGCTGGACGCTGTCATCGAGGCGGAGGCTCTGCTCCGCGAGCTGGGCTGTCTGGACGCCGAGGACGCGCTCACGCCACTCGGGACCATCCTCGCCAAGCTGCCGATAGAACCTCGCCTCGGCAAAATGATGGTGTTAGGCTTTGTGCTTGGCGTTGGAGACGCCCTGACCACGATGGCGGCTAATTCTACAACGTTCCCGGAAATATTCGTGCTCGAAGGCCGGAGGAGACTGTCGATGCACCAGAGAGCCCTGGGCGGAGACCGGGCCTCCGACCACGTGGCCATGTTGAATGCATTCCAGATGTGGGAGAGAGAACACAACAAAGGCGAAGAGGCAGAGCTACGGTGGTGCGAATGGAAGGGCGTACAGCAGACCACGCTGCGTGTTACTTACGAGGCTAAACATCAATTGATAAATATCCTCACGACCGCAATAGGCTTCAACGAAGAATGCTGTGTTCCACAACGTTGGATGCCGAACGGACCGGACCCAACTTTGGACTTGGTCATAGCCCTCATGTGCATGGGCCTGTACCCTAACGTTTGTCTACACCAAGGGAAGAGGAAGGTATTAACGACGGAAGGGAAACCGGCATTGATACACAAGACCTCAGTGAACTGCAGCAACATGGAGCAGAAGTTCCCCTCGCCTCTGTTCGTTTTCGGCGAGAAGGTGCGCACCCGGGCCATCAGCTGCAAGCAGACCACCATGGTCGCTCCCATACATCTGCTGCTCTTCGGCTGTAACAAAGTGGAATGGGTGGACAACGTGGTGCGGCTGGACAACTGGCTGAACTTCCAGATGTCGCCGCGATCGGCCGCCCTCATCATAGCGCTGCGGCCCGCCATCGAGAGGATCGTGGAGCGCGCGGCGGCCCAACCCGACGCGGCGCTGCAGTTCTCGCCGGCAGAGCGAAAGGTGGTAGAATGTCTGCGGGAGCTTTGTGTGACGGACGCCGGTGACTACAACATAAAACGAGACGTCAACATGCCATCGTTCCGATCCGAAGGTCCAAAACGATCCATGAGAGGATGGGGAACTAACGGACCTCGAAGTGGTTTTGGTGGTGGAGGGACCACAGGCAGAGGCTTTACCCAAGGATTAGGAAACCAAGGTGCTTTCGGGACGCGCTTCGGAAATGGTGGTATCAGCACTCAAAGCAGTCAAGGCGGTTTTTGGAATCAAGGTGGCAACTTTGGCGGAAGAGGCGATTTTAATAGCGGCCGTGGAAGTTTTGGTAACCAAGGAAGCTATGGTAACCAAGGAAGCTATGGTAACCAAGGAAGCTATGGTAACCAAGGAAGCTTCGGTAACCAAGGTAGCTTTGGTAACCAAGGAAGCTTGGGTAACCATGGAAGCTTTGGTAGTTCTAGGGGATCCGGGTTCGGTAACAGAAATCAGGGAAACTCTAGTTGGACACAACGTTATTAA
Protein
MDIKSFLYTWCAKKGLTPLFDIRATGPKHRQRFLCEVRVEGFSYLGAGNSTTKKDAQMNAAKDFVSYLVRAGQIPQADVPEDVKAKAIEESTNEESVENNGGQQPKQVFQDGMGPEVIGPAYQAYGGENDKKFSYIDRMQQQKNVEEAEDLDINSSLHGNWTMENAKSKLHQFLQMNKINADYVYKAVGPDHTRSFACELTIYVSQLGRNVTGRETASNKQTASKSCALSIVRQLYHLGVIEAYSGTLKRDRSSEGIMKPYPVAICPELEQQLEDTLQELEIEPVNVDAPSQDNQEGSAVTLLQKDRAKAMREAKPMPAGVVTWSPPQPNWNAWTGCNIDEGALATASMDEISADLEKCHRNICQNSKLYQESLNEREHLPVFSMKSQIMEAINENPVIIIRGNTGCGKTTQVCQFILDDYIASGQGAWANVCVTQPRRISAVSVAERVAAERCEELGNTVGYSVRFESVLPRPYGSIMFCTVGVLLRKLEGGLRGVSHVLVDEVHERDADTDFALILLRDMAHTYPDLRIILMSATVDTTLFVKYFGGCPVIEVPGRTYPVTQYFLEDSIELTKFMPPPITRKRKSTGKRANKDDEDDDDEDDLDEPYEDLNKQCSLGDGYSQATVDALQQLSERDFSFELVQAILMYIDGLGGDGAVLVFLPGWNLIFALMKHLLQHRLFGDPSKYVILPLHSQIPREDQKKVFITPPEGITKVILSTNIAETSITINDVVYVIDSCKAKMKLFTSHNNMTSYATVWASKTNLEQRKGRAGRVRPGVCFTLCTYARYEKLEEHLAAEMFRTPLHELALSIKLLRLGAIGHFLSKAPEPPPLDAVIEAEALLRELGCLDAEDALTPLGTILAKLPIEPRLGKMMVLGFVLGVGDALTTMAANSTTFPEIFVLEGRRRLSMHQRALGGDRASDHVAMLNAFQMWEREHNKGEEAELRWCEWKGVQQTTLRVTYEAKHQLINILTTAIGFNEECCVPQRWMPNGPDPTLDLVIALMCMGLYPNVCLHQGKRKVLTTEGKPALIHKTSVNCSNMEQKFPSPLFVFGEKVRTRAISCKQTTMVAPIHLLLFGCNKVEWVDNVVRLDNWLNFQMSPRSAALIIALRPAIERIVERAAAQPDAALQFSPAERKVVECLRELCVTDAGDYNIKRDVNMPSFRSEGPKRSMRGWGTNGPRSGFGGGGTTGRGFTQGLGNQGAFGTRFGNGGISTQSSQGGFWNQGGNFGGRGDFNSGRGSFGNQGSYGNQGSYGNQGSYGNQGSFGNQGSFGNQGSLGNHGSFGSSRGSGFGNRNQGNSSWTQRY
Summary
Description
Required in males for dosage compensation of X chromosome linked genes. Mle, msl-1 and msl-3 are colocalized on X chromosome. Each of the msl proteins requires all the other msls for wild-type X-chromosome binding. Probably unwinds double-stranded DNA and RNA in a 3' to 5' direction.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Interacts with Top2.
Similarity
Belongs to the DEAD box helicase family. DEAH subfamily.
Keywords
3D-structure
Alternative splicing
ATP-binding
Chromosome
Complete proteome
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Reference proteome
Repeat
Feature
chain Dosage compensation regulator
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A5JPM0
H9JMT7
A0A194PL29
A0A0E3T0T7
A0A1W4XQL2
A0A2P8Y1S8
+ More
A0A0N0U6F1 E2AJX4 A0A1Y1MYC3 A0A154PBM7 D6WEI4 A0A195FJ67 A0A195E515 A0A067REK5 A0A026WEH8 A0A195BT45 A0A158P2N2 A0A0L7R9U8 A0A310SAZ9 A0A0C9RMC0 A0A195D312 A0A151X2S4 E2C021 A0A2J7R184 A0A087ZRK5 A0A232F4E4 K7J0N6 A0A224X5G5 A0A023F369 A0A069DY95 F4W6K4 A0A2A3ENZ3 A0A1B6BXD9 A0A146LDN6 U5EXL1 A0A0L0CKH7 T1PI79 O77403 A0A336M9P6 A0A1I8MSX9 A0A1I8P1P2 A0A0K8V382 U4UXZ4 E0W3Q0 W8BUA8 A0A1J1ILZ8 A0A0A1X440 N6UDI4 A0A1A9ZS37 A0A1B0FDM8 A0A1A9WLJ1 A0A1A9V464 A0A182GDT3 A0A3R5WVZ0 B3N3H1 B3STH3 B3STH0 B3STG2 B3STG6 P24785 B3STG3 B3STH6 B3STI2 B3STG7 B4QCQ3 B3STI1 B3STG9 B3STH2 B3STG5 A0A182WBR6 B3STH7 B3STH8 B3STH9 A0A182VNR7 A0A182M585 B4II22 B3STG4 A0A182UEC7 B3STI0 B3STI3 Q7Q577 A0A182X4I0 B4NYW3 A0A3B0J7F0 A0A182I4H7 A0A182R947 A0A182QSZ8 A0A182Y2K8 A0A182NIA7 Q28YQ6 A0A182JYZ5 A8ILB3 B4GJ01 B3MJ84 A0A087TNK7 B4MDY4 A0A1W4V2D0 A8IL99 A0A1A9YEF3 A0A182JE01
A0A0N0U6F1 E2AJX4 A0A1Y1MYC3 A0A154PBM7 D6WEI4 A0A195FJ67 A0A195E515 A0A067REK5 A0A026WEH8 A0A195BT45 A0A158P2N2 A0A0L7R9U8 A0A310SAZ9 A0A0C9RMC0 A0A195D312 A0A151X2S4 E2C021 A0A2J7R184 A0A087ZRK5 A0A232F4E4 K7J0N6 A0A224X5G5 A0A023F369 A0A069DY95 F4W6K4 A0A2A3ENZ3 A0A1B6BXD9 A0A146LDN6 U5EXL1 A0A0L0CKH7 T1PI79 O77403 A0A336M9P6 A0A1I8MSX9 A0A1I8P1P2 A0A0K8V382 U4UXZ4 E0W3Q0 W8BUA8 A0A1J1ILZ8 A0A0A1X440 N6UDI4 A0A1A9ZS37 A0A1B0FDM8 A0A1A9WLJ1 A0A1A9V464 A0A182GDT3 A0A3R5WVZ0 B3N3H1 B3STH3 B3STH0 B3STG2 B3STG6 P24785 B3STG3 B3STH6 B3STI2 B3STG7 B4QCQ3 B3STI1 B3STG9 B3STH2 B3STG5 A0A182WBR6 B3STH7 B3STH8 B3STH9 A0A182VNR7 A0A182M585 B4II22 B3STG4 A0A182UEC7 B3STI0 B3STI3 Q7Q577 A0A182X4I0 B4NYW3 A0A3B0J7F0 A0A182I4H7 A0A182R947 A0A182QSZ8 A0A182Y2K8 A0A182NIA7 Q28YQ6 A0A182JYZ5 A8ILB3 B4GJ01 B3MJ84 A0A087TNK7 B4MDY4 A0A1W4V2D0 A8IL99 A0A1A9YEF3 A0A182JE01
EC Number
3.6.4.13
Pubmed
19121390
26354079
29403074
20798317
28004739
18362917
+ More
19820115 24845553 24508170 21347285 28648823 20075255 25474469 26334808 21719571 26823975 26108605 11102379 25315136 23537049 20566863 24495485 25830018 26483478 17994087 1653648 10731132 12537572 23989663 22936249 12364791 17550304 25244985 15632085 18039888
19820115 24845553 24508170 21347285 28648823 20075255 25474469 26334808 21719571 26823975 26108605 11102379 25315136 23537049 20566863 24495485 25830018 26483478 17994087 1653648 10731132 12537572 23989663 22936249 12364791 17550304 25244985 15632085 18039888
EMBL
EF554693
ABQ51917.1
BABH01016861
BABH01016862
BABH01016863
KQ459601
+ More
KPI93703.1 KP236739 AKB96195.1 PYGN01001041 PSN38206.1 KQ435742 KOX76701.1 GL440100 EFN66290.1 GEZM01017457 JAV90663.1 KQ434869 KZC09251.1 KQ971318 EFA00345.2 KQ981523 KYN40307.1 KQ979657 KYN19949.1 KK852517 KDR22182.1 KK107250 EZA54348.1 KQ976408 KYM91133.1 ADTU01007051 KQ414625 KOC67531.1 KQ762031 OAD56377.1 GBYB01009460 JAG79227.1 KQ976885 KYN07300.1 KQ982580 KYQ54504.1 GL451712 EFN78717.1 NEVH01008213 PNF34584.1 NNAY01001035 OXU25399.1 GFTR01008689 JAW07737.1 GBBI01003111 JAC15601.1 GBGD01000152 JAC88737.1 GL887707 EGI70339.1 KZ288202 PBC33485.1 GEDC01031628 JAS05670.1 GDHC01012880 JAQ05749.1 GANO01002490 JAB57381.1 JRES01000534 JRES01000356 KNC30377.1 KNC31969.1 KA647845 AFP62474.1 Y18119 CAA77038.1 UFQT01000602 SSX25629.1 GDHF01019259 JAI33055.1 KB632411 ERL95241.1 DS235882 EEB20256.1 GAMC01003773 JAC02783.1 CVRI01000054 CRL00094.1 GBXI01008859 JAD05433.1 APGK01032721 APGK01032722 KB740848 ENN78686.1 CCAG010004130 JXUM01016703 MG708324 QAB02864.1 CH954177 EDV59853.1 EF682050 ABV48872.1 EF682047 EF682048 EF682051 ABV48869.1 ABV48870.1 ABV48873.1 EF682039 ABV48861.1 EF682043 EF682045 EF682052 JQ663522 ABV48865.1 ABV48867.1 ABV48874.1 AFI26242.1 M74121 AE013599 BT003785 BT010267 EF682040 ABV48862.1 EF682053 ABV48875.1 EF682059 ABV48881.1 EF682044 ABV48866.1 CM000362 CM002911 EDX05842.1 KMY91671.1 EF682058 ABV48880.1 EF682046 ABV48868.1 EF682049 ABV48871.1 EF682042 ABV48864.1 EF682054 ABV48876.1 EF682055 ABV48877.1 EF682056 ABV48878.1 AXCM01000119 CH480841 EDW49548.1 EF682041 ABV48863.1 EF682057 ABV48879.1 EF682060 ABV48882.1 AAAB01008960 EAA11305.4 CM000157 EDW89814.1 OUUW01000001 SPP75832.1 APCN01002313 AXCN02000879 CM000071 EAL25909.2 EU167133 ABV82525.1 CH479183 EDW36419.1 CH902619 EDV38178.1 KK116061 KFM66696.1 CH940662 EDW58749.2 EU167129 ABV82521.1
KPI93703.1 KP236739 AKB96195.1 PYGN01001041 PSN38206.1 KQ435742 KOX76701.1 GL440100 EFN66290.1 GEZM01017457 JAV90663.1 KQ434869 KZC09251.1 KQ971318 EFA00345.2 KQ981523 KYN40307.1 KQ979657 KYN19949.1 KK852517 KDR22182.1 KK107250 EZA54348.1 KQ976408 KYM91133.1 ADTU01007051 KQ414625 KOC67531.1 KQ762031 OAD56377.1 GBYB01009460 JAG79227.1 KQ976885 KYN07300.1 KQ982580 KYQ54504.1 GL451712 EFN78717.1 NEVH01008213 PNF34584.1 NNAY01001035 OXU25399.1 GFTR01008689 JAW07737.1 GBBI01003111 JAC15601.1 GBGD01000152 JAC88737.1 GL887707 EGI70339.1 KZ288202 PBC33485.1 GEDC01031628 JAS05670.1 GDHC01012880 JAQ05749.1 GANO01002490 JAB57381.1 JRES01000534 JRES01000356 KNC30377.1 KNC31969.1 KA647845 AFP62474.1 Y18119 CAA77038.1 UFQT01000602 SSX25629.1 GDHF01019259 JAI33055.1 KB632411 ERL95241.1 DS235882 EEB20256.1 GAMC01003773 JAC02783.1 CVRI01000054 CRL00094.1 GBXI01008859 JAD05433.1 APGK01032721 APGK01032722 KB740848 ENN78686.1 CCAG010004130 JXUM01016703 MG708324 QAB02864.1 CH954177 EDV59853.1 EF682050 ABV48872.1 EF682047 EF682048 EF682051 ABV48869.1 ABV48870.1 ABV48873.1 EF682039 ABV48861.1 EF682043 EF682045 EF682052 JQ663522 ABV48865.1 ABV48867.1 ABV48874.1 AFI26242.1 M74121 AE013599 BT003785 BT010267 EF682040 ABV48862.1 EF682053 ABV48875.1 EF682059 ABV48881.1 EF682044 ABV48866.1 CM000362 CM002911 EDX05842.1 KMY91671.1 EF682058 ABV48880.1 EF682046 ABV48868.1 EF682049 ABV48871.1 EF682042 ABV48864.1 EF682054 ABV48876.1 EF682055 ABV48877.1 EF682056 ABV48878.1 AXCM01000119 CH480841 EDW49548.1 EF682041 ABV48863.1 EF682057 ABV48879.1 EF682060 ABV48882.1 AAAB01008960 EAA11305.4 CM000157 EDW89814.1 OUUW01000001 SPP75832.1 APCN01002313 AXCN02000879 CM000071 EAL25909.2 EU167133 ABV82525.1 CH479183 EDW36419.1 CH902619 EDV38178.1 KK116061 KFM66696.1 CH940662 EDW58749.2 EU167129 ABV82521.1
Proteomes
UP000005204
UP000053268
UP000192223
UP000245037
UP000053105
UP000000311
+ More
UP000076502 UP000007266 UP000078541 UP000078492 UP000027135 UP000053097 UP000078540 UP000005205 UP000053825 UP000078542 UP000075809 UP000008237 UP000235965 UP000005203 UP000215335 UP000002358 UP000007755 UP000242457 UP000037069 UP000095301 UP000095300 UP000030742 UP000009046 UP000183832 UP000019118 UP000092445 UP000092444 UP000091820 UP000078200 UP000069940 UP000008711 UP000000803 UP000000304 UP000075920 UP000075903 UP000075883 UP000001292 UP000075902 UP000007062 UP000076407 UP000002282 UP000268350 UP000075840 UP000075900 UP000075886 UP000076408 UP000075884 UP000001819 UP000075881 UP000008744 UP000007801 UP000054359 UP000008792 UP000192221 UP000092443 UP000075880
UP000076502 UP000007266 UP000078541 UP000078492 UP000027135 UP000053097 UP000078540 UP000005205 UP000053825 UP000078542 UP000075809 UP000008237 UP000235965 UP000005203 UP000215335 UP000002358 UP000007755 UP000242457 UP000037069 UP000095301 UP000095300 UP000030742 UP000009046 UP000183832 UP000019118 UP000092445 UP000092444 UP000091820 UP000078200 UP000069940 UP000008711 UP000000803 UP000000304 UP000075920 UP000075903 UP000075883 UP000001292 UP000075902 UP000007062 UP000076407 UP000002282 UP000268350 UP000075840 UP000075900 UP000075886 UP000076408 UP000075884 UP000001819 UP000075881 UP000008744 UP000007801 UP000054359 UP000008792 UP000192221 UP000092443 UP000075880
Pfam
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A5JPM0
H9JMT7
A0A194PL29
A0A0E3T0T7
A0A1W4XQL2
A0A2P8Y1S8
+ More
A0A0N0U6F1 E2AJX4 A0A1Y1MYC3 A0A154PBM7 D6WEI4 A0A195FJ67 A0A195E515 A0A067REK5 A0A026WEH8 A0A195BT45 A0A158P2N2 A0A0L7R9U8 A0A310SAZ9 A0A0C9RMC0 A0A195D312 A0A151X2S4 E2C021 A0A2J7R184 A0A087ZRK5 A0A232F4E4 K7J0N6 A0A224X5G5 A0A023F369 A0A069DY95 F4W6K4 A0A2A3ENZ3 A0A1B6BXD9 A0A146LDN6 U5EXL1 A0A0L0CKH7 T1PI79 O77403 A0A336M9P6 A0A1I8MSX9 A0A1I8P1P2 A0A0K8V382 U4UXZ4 E0W3Q0 W8BUA8 A0A1J1ILZ8 A0A0A1X440 N6UDI4 A0A1A9ZS37 A0A1B0FDM8 A0A1A9WLJ1 A0A1A9V464 A0A182GDT3 A0A3R5WVZ0 B3N3H1 B3STH3 B3STH0 B3STG2 B3STG6 P24785 B3STG3 B3STH6 B3STI2 B3STG7 B4QCQ3 B3STI1 B3STG9 B3STH2 B3STG5 A0A182WBR6 B3STH7 B3STH8 B3STH9 A0A182VNR7 A0A182M585 B4II22 B3STG4 A0A182UEC7 B3STI0 B3STI3 Q7Q577 A0A182X4I0 B4NYW3 A0A3B0J7F0 A0A182I4H7 A0A182R947 A0A182QSZ8 A0A182Y2K8 A0A182NIA7 Q28YQ6 A0A182JYZ5 A8ILB3 B4GJ01 B3MJ84 A0A087TNK7 B4MDY4 A0A1W4V2D0 A8IL99 A0A1A9YEF3 A0A182JE01
A0A0N0U6F1 E2AJX4 A0A1Y1MYC3 A0A154PBM7 D6WEI4 A0A195FJ67 A0A195E515 A0A067REK5 A0A026WEH8 A0A195BT45 A0A158P2N2 A0A0L7R9U8 A0A310SAZ9 A0A0C9RMC0 A0A195D312 A0A151X2S4 E2C021 A0A2J7R184 A0A087ZRK5 A0A232F4E4 K7J0N6 A0A224X5G5 A0A023F369 A0A069DY95 F4W6K4 A0A2A3ENZ3 A0A1B6BXD9 A0A146LDN6 U5EXL1 A0A0L0CKH7 T1PI79 O77403 A0A336M9P6 A0A1I8MSX9 A0A1I8P1P2 A0A0K8V382 U4UXZ4 E0W3Q0 W8BUA8 A0A1J1ILZ8 A0A0A1X440 N6UDI4 A0A1A9ZS37 A0A1B0FDM8 A0A1A9WLJ1 A0A1A9V464 A0A182GDT3 A0A3R5WVZ0 B3N3H1 B3STH3 B3STH0 B3STG2 B3STG6 P24785 B3STG3 B3STH6 B3STI2 B3STG7 B4QCQ3 B3STI1 B3STG9 B3STH2 B3STG5 A0A182WBR6 B3STH7 B3STH8 B3STH9 A0A182VNR7 A0A182M585 B4II22 B3STG4 A0A182UEC7 B3STI0 B3STI3 Q7Q577 A0A182X4I0 B4NYW3 A0A3B0J7F0 A0A182I4H7 A0A182R947 A0A182QSZ8 A0A182Y2K8 A0A182NIA7 Q28YQ6 A0A182JYZ5 A8ILB3 B4GJ01 B3MJ84 A0A087TNK7 B4MDY4 A0A1W4V2D0 A8IL99 A0A1A9YEF3 A0A182JE01
PDB
5AOR
E-value=0,
Score=3437
Ontologies
GO
GO:0005524
GO:0004386
GO:0003723
GO:0050684
GO:1990904
GO:0045944
GO:0043140
GO:0005634
GO:0005730
GO:0004003
GO:0034459
GO:0016021
GO:0016787
GO:0007549
GO:0072487
GO:0048675
GO:0000785
GO:0016887
GO:0031453
GO:0005829
GO:0008340
GO:0001069
GO:0003682
GO:0005700
GO:0000228
GO:0000805
GO:0045433
GO:0003690
GO:0032508
GO:2000765
GO:0005694
GO:0009047
GO:0016456
GO:0016457
GO:2000373
GO:0003724
GO:0008026
GO:0003725
GO:0003676
GO:0030126
GO:0030117
GO:0006418
GO:0003707
GO:0006281
GO:0006259
Topology
Subcellular location
Nucleus
Chromosome
Chromosome
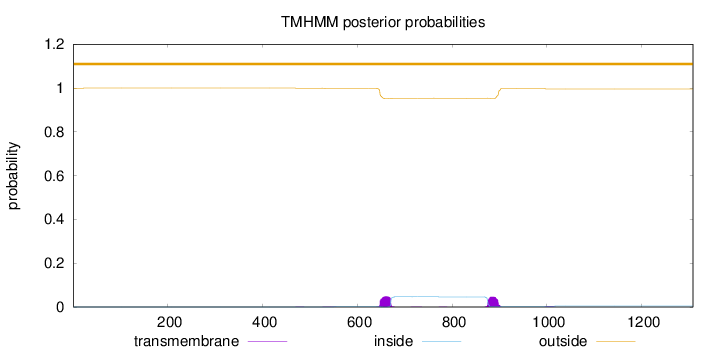
Length:
1308
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.19370000000001
Exp number, first 60 AAs:
0.00192
Total prob of N-in:
0.00040
outside
1 - 1308
Population Genetic Test Statistics
Pi
232.504595
Theta
196.718611
Tajima's D
0.963932
CLR
0.08586
CSRT
0.653917304134793
Interpretation
Uncertain
