Pre Gene Modal
BGIBMGA010839
Annotation
PREDICTED:_vesicular_integral-membrane_protein_VIP36_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.08
Sequence
CDS
ATGAAGTTGGACTTTTCAACAATGATTCATCAGAGAATAAGCTGGGTGCAAATCCTTTGCGCTACGTTATTTCTCAACCCAATATTTGCAGAATGGAATACAAGGGACTACATAAGAAGAGAGCATTCACTGACAAAACCCTATCAAGGTAGTGGAATGTCCTTGCCATATTGGGATTTTTTAGGTAGTACAATTGTAACCTCGAATTATGTGCGTCTAACACCCGATTTGCAATCAAAAGCTGGTGCTCTATGGAATACTGTTCCCTGTAACACTAGAAATTGGGAACTGCAGGTGCAATTCAAAGTGCATGGTCGAGGAAAAGAGCTATTTGGTGATGGATTTGCAATATGGTATGTCAAAGAGAGAATGGAAAGTGGCCCTGTATTTGGCAGTAAGGACTACTTCAATGGCTTAGCAATCATACTCGACACATACAGCAACCACAATGGTGCTCATAATCACCAGCATCCATACATATCAGCAATGGTAAATAATGGAACACTCCACTACGACCACGATCGTGATGGAACCCACACTCAGCTCTCTGGCTGCGAAGCAAAGTTTCGAAATTTCAACCATGATTCACATATTTCTATAGTGTATAGGGATGATACACTTATTGTATCAACTGATTTGGAAGGCAAGAATGCCTGGAAGGAATGTTTGAAAGTGGAGAATGTCCTGTTGCCTACTGGTTACTACTTTGGTGCCTCTGCCACTACTGGAGACTTAAGTGATAATCATGACATTATCTCTATAAAAATGTTTGAACTTGATCTACTTGAATCTCAAAAAAAAGACGAAGATAGATCTCACATTATCCCTTCAGCAGCCTCATTCGAAGCACCACGCGAACGCATCGAAGACGCCAAGCCAGCCATGTCAGGCATCAAAACATTCCTCTGGATGATGTTTGTTGCCATTGTAATCATTGTGCTAGTCGTTCTCGGTATCATGTGGTACCAAAAACGCCAAGAGCAGTCCAGAAAAAGACTTTATTGA
Protein
MKLDFSTMIHQRISWVQILCATLFLNPIFAEWNTRDYIRREHSLTKPYQGSGMSLPYWDFLGSTIVTSNYVRLTPDLQSKAGALWNTVPCNTRNWELQVQFKVHGRGKELFGDGFAIWYVKERMESGPVFGSKDYFNGLAIILDTYSNHNGAHNHQHPYISAMVNNGTLHYDHDRDGTHTQLSGCEAKFRNFNHDSHISIVYRDDTLIVSTDLEGKNAWKECLKVENVLLPTGYYFGASATTGDLSDNHDIISIKMFELDLLESQKKDEDRSHIIPSAASFEAPRERIEDAKPAMSGIKTFLWMMFVAIVIIVLVVLGIMWYQKRQEQSRKRLY
Summary
Uniprot
H9JMT6
A0A2A4JCQ2
E7EC18
A0A0L7LRV7
A0A2H1WIV8
A0A212EM81
+ More
A0A1E1WTZ7 S4PCC9 A0A194PJU8 E2BH30 K7J8S0 E9ICB0 A0A067R7B1 A0A154PSX6 V9IFA2 A0A151WQX0 A0A088AEZ9 A0A151I8V2 A0A026WBZ9 A0A0L7QKW1 A0A0M8ZUN1 A0A158NXN3 A0A232EYZ5 A0A1B6FSF5 A0A1Y1KZI7 D6X3B0 A0A1B6LHU9 A0A2A3ECC8 A0A1B6IB06 A0A195BRR8 A0A0N8CR05 A0A0P4ZPL0 A0A0P5CR21 J3JZI3 A0A0N7ZPL1 A0A195E4M0 B7QFT9 A0A1B6CZ37 A0A0T6B360 A0A0K8RMH6 E0VUZ8 E9G8W7 A0A293MRX8 A0A0P6AH22 A0A182XF66 Q7QA71 A0A182HVN7 A0A182V8Q8 A0A182KZS1 A0A2R5L9I5 A0A182PLG9 A0A182UAE7 V5IC29 A0A1L8E3B8 U4UM14 A0A182Y909 A0A1L8E362 A0A1L8E2V9 A0A1L8E2X2 N6SYS6 A0A182RI26 A0A182TBZ5 A0A182FAF3 A0A2M3ZKW9 A0A2M4AKU1 A0A2M4BTM6 A0A182MT70 A0A0P5WGH0 T1DNL0 W5JWA7 A0A0P5XHL4 A0A084VDK1 A0A182W383 A0A0P6JRH1 A0A0P5P1W9 A0A1B0GI93 A0A182K4W2 T1ISH8 Q16SI7 A0A224Z396 A0A131Z571 A0A023FYY7 L7LYV5 A0A0P6BXR7 G3MI02 A0A0A9VTR0 A0A182ILX8 A0A023FPA0 A0A182GZZ9 U5ER85 A0A023FER3 T1E2S8 A0A1Z5LBH1 B0W3A9 A0A1Q3F5W4 A0A1Q3F605 B4MBX6 A0A0N8ECK5 A0A0P5TUB7 A0A087TWC9
A0A1E1WTZ7 S4PCC9 A0A194PJU8 E2BH30 K7J8S0 E9ICB0 A0A067R7B1 A0A154PSX6 V9IFA2 A0A151WQX0 A0A088AEZ9 A0A151I8V2 A0A026WBZ9 A0A0L7QKW1 A0A0M8ZUN1 A0A158NXN3 A0A232EYZ5 A0A1B6FSF5 A0A1Y1KZI7 D6X3B0 A0A1B6LHU9 A0A2A3ECC8 A0A1B6IB06 A0A195BRR8 A0A0N8CR05 A0A0P4ZPL0 A0A0P5CR21 J3JZI3 A0A0N7ZPL1 A0A195E4M0 B7QFT9 A0A1B6CZ37 A0A0T6B360 A0A0K8RMH6 E0VUZ8 E9G8W7 A0A293MRX8 A0A0P6AH22 A0A182XF66 Q7QA71 A0A182HVN7 A0A182V8Q8 A0A182KZS1 A0A2R5L9I5 A0A182PLG9 A0A182UAE7 V5IC29 A0A1L8E3B8 U4UM14 A0A182Y909 A0A1L8E362 A0A1L8E2V9 A0A1L8E2X2 N6SYS6 A0A182RI26 A0A182TBZ5 A0A182FAF3 A0A2M3ZKW9 A0A2M4AKU1 A0A2M4BTM6 A0A182MT70 A0A0P5WGH0 T1DNL0 W5JWA7 A0A0P5XHL4 A0A084VDK1 A0A182W383 A0A0P6JRH1 A0A0P5P1W9 A0A1B0GI93 A0A182K4W2 T1ISH8 Q16SI7 A0A224Z396 A0A131Z571 A0A023FYY7 L7LYV5 A0A0P6BXR7 G3MI02 A0A0A9VTR0 A0A182ILX8 A0A023FPA0 A0A182GZZ9 U5ER85 A0A023FER3 T1E2S8 A0A1Z5LBH1 B0W3A9 A0A1Q3F5W4 A0A1Q3F605 B4MBX6 A0A0N8ECK5 A0A0P5TUB7 A0A087TWC9
Pubmed
19121390
26227816
22118469
23622113
26354079
20798317
+ More
20075255 21282665 24845553 24508170 30249741 21347285 28648823 28004739 18362917 19820115 22516182 20566863 21292972 12364791 14747013 17210077 20966253 25765539 23537049 25244985 20920257 23761445 24438588 17510324 28797301 26830274 25576852 22216098 25401762 26823975 26483478 24330624 28528879 17994087
20075255 21282665 24845553 24508170 30249741 21347285 28648823 28004739 18362917 19820115 22516182 20566863 21292972 12364791 14747013 17210077 20966253 25765539 23537049 25244985 20920257 23761445 24438588 17510324 28797301 26830274 25576852 22216098 25401762 26823975 26483478 24330624 28528879 17994087
EMBL
BABH01016860
NWSH01002104
PCG69173.1
HQ645942
ADU25045.1
JTDY01000233
+ More
KOB78139.1 ODYU01008904 SOQ52896.1 AGBW02013887 OWR42588.1 GDQN01000610 JAT90444.1 GAIX01004241 JAA88319.1 KQ459601 KPI93706.1 GL448268 EFN84951.1 GL762231 EFZ21795.1 KK852895 KDR14191.1 KQ435180 KZC15011.1 JR041046 AEY59347.1 KQ982815 KYQ50240.1 KQ978326 KYM95094.1 KK107323 QOIP01000002 EZA52584.1 RLU25875.1 KQ414940 KOC59220.1 KQ435879 KOX69869.1 ADTU01000475 ADTU01000476 NNAY01001520 OXU23706.1 GECZ01016651 JAS53118.1 GEZM01075069 JAV64307.1 KQ971372 EFA10355.1 GEBQ01016758 JAT23219.1 KZ288287 PBC29433.1 GECU01023609 JAS84097.1 KQ976423 KYM88919.1 GDIP01107407 JAL96307.1 GDIP01214397 GDIP01210645 GDIQ01040027 LRGB01002451 JAJ12757.1 JAN54710.1 KZS07650.1 GDIP01166736 JAJ56666.1 BT128665 AEE63622.1 GDIP01225049 JAI98352.1 KQ979685 KYN19804.1 ABJB010313462 ABJB010369059 ABJB010817547 ABJB010859285 ABJB011110643 DS927877 EEC17711.1 GEDC01018765 GEDC01005033 JAS18533.1 JAS32265.1 LJIG01016044 KRT81782.1 GADI01001710 JAA72098.1 DS235797 EEB17204.1 GL732535 EFX84199.1 GFWV01018824 MAA43552.1 GDIP01042770 JAM60945.1 AAAB01008898 EAA09126.4 APCN01005808 GGLE01002012 MBY06138.1 GANP01014407 JAB70061.1 GFDF01001012 JAV13072.1 KB632271 ERL91045.1 GFDF01001005 JAV13079.1 GFDF01001004 JAV13080.1 GFDF01001013 JAV13071.1 APGK01059049 KB741292 ENN70383.1 GGFM01008392 MBW29143.1 GGFK01008082 MBW41403.1 GGFJ01007223 MBW56364.1 AXCM01000104 GDIP01099611 JAM04104.1 GAMD01002760 JAA98830.1 ADMH02000180 ETN67460.1 GDIP01071966 JAM31749.1 ATLV01011556 KE524695 KFB36045.1 GDIQ01011422 JAN83315.1 GDIQ01134693 JAL17033.1 AJWK01013575 JH431432 CH477673 EAT37418.1 GFPF01009416 MAA20562.1 GEDV01002100 JAP86457.1 GBBL01001410 JAC25910.1 GACK01008237 JAA56797.1 GDIP01009090 JAM94625.1 JO841503 AEO33120.1 GBHO01045013 GBHO01017606 GBRD01001718 GDHC01005497 JAF98590.1 JAG25998.1 JAG64103.1 JAQ13132.1 GBBK01001797 JAC22685.1 JXUM01100343 JXUM01100344 KQ564561 KXJ72087.1 GANO01002916 JAB56955.1 GBBK01004500 JAC19982.1 GALA01001025 JAA93827.1 GFJQ02002402 JAW04568.1 DS231831 EDS31252.1 GFDL01012142 JAV22903.1 GFDL01012102 JAV22943.1 CH940656 EDW58597.1 GDIQ01040028 JAN54709.1 GDIP01121502 JAL82212.1 KK117063 KFM69418.1
KOB78139.1 ODYU01008904 SOQ52896.1 AGBW02013887 OWR42588.1 GDQN01000610 JAT90444.1 GAIX01004241 JAA88319.1 KQ459601 KPI93706.1 GL448268 EFN84951.1 GL762231 EFZ21795.1 KK852895 KDR14191.1 KQ435180 KZC15011.1 JR041046 AEY59347.1 KQ982815 KYQ50240.1 KQ978326 KYM95094.1 KK107323 QOIP01000002 EZA52584.1 RLU25875.1 KQ414940 KOC59220.1 KQ435879 KOX69869.1 ADTU01000475 ADTU01000476 NNAY01001520 OXU23706.1 GECZ01016651 JAS53118.1 GEZM01075069 JAV64307.1 KQ971372 EFA10355.1 GEBQ01016758 JAT23219.1 KZ288287 PBC29433.1 GECU01023609 JAS84097.1 KQ976423 KYM88919.1 GDIP01107407 JAL96307.1 GDIP01214397 GDIP01210645 GDIQ01040027 LRGB01002451 JAJ12757.1 JAN54710.1 KZS07650.1 GDIP01166736 JAJ56666.1 BT128665 AEE63622.1 GDIP01225049 JAI98352.1 KQ979685 KYN19804.1 ABJB010313462 ABJB010369059 ABJB010817547 ABJB010859285 ABJB011110643 DS927877 EEC17711.1 GEDC01018765 GEDC01005033 JAS18533.1 JAS32265.1 LJIG01016044 KRT81782.1 GADI01001710 JAA72098.1 DS235797 EEB17204.1 GL732535 EFX84199.1 GFWV01018824 MAA43552.1 GDIP01042770 JAM60945.1 AAAB01008898 EAA09126.4 APCN01005808 GGLE01002012 MBY06138.1 GANP01014407 JAB70061.1 GFDF01001012 JAV13072.1 KB632271 ERL91045.1 GFDF01001005 JAV13079.1 GFDF01001004 JAV13080.1 GFDF01001013 JAV13071.1 APGK01059049 KB741292 ENN70383.1 GGFM01008392 MBW29143.1 GGFK01008082 MBW41403.1 GGFJ01007223 MBW56364.1 AXCM01000104 GDIP01099611 JAM04104.1 GAMD01002760 JAA98830.1 ADMH02000180 ETN67460.1 GDIP01071966 JAM31749.1 ATLV01011556 KE524695 KFB36045.1 GDIQ01011422 JAN83315.1 GDIQ01134693 JAL17033.1 AJWK01013575 JH431432 CH477673 EAT37418.1 GFPF01009416 MAA20562.1 GEDV01002100 JAP86457.1 GBBL01001410 JAC25910.1 GACK01008237 JAA56797.1 GDIP01009090 JAM94625.1 JO841503 AEO33120.1 GBHO01045013 GBHO01017606 GBRD01001718 GDHC01005497 JAF98590.1 JAG25998.1 JAG64103.1 JAQ13132.1 GBBK01001797 JAC22685.1 JXUM01100343 JXUM01100344 KQ564561 KXJ72087.1 GANO01002916 JAB56955.1 GBBK01004500 JAC19982.1 GALA01001025 JAA93827.1 GFJQ02002402 JAW04568.1 DS231831 EDS31252.1 GFDL01012142 JAV22903.1 GFDL01012102 JAV22943.1 CH940656 EDW58597.1 GDIQ01040028 JAN54709.1 GDIP01121502 JAL82212.1 KK117063 KFM69418.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000008237
+ More
UP000002358 UP000027135 UP000076502 UP000075809 UP000005203 UP000078542 UP000053097 UP000279307 UP000053825 UP000053105 UP000005205 UP000215335 UP000007266 UP000242457 UP000078540 UP000076858 UP000078492 UP000001555 UP000009046 UP000000305 UP000076407 UP000007062 UP000075840 UP000075903 UP000075882 UP000075885 UP000075902 UP000030742 UP000076408 UP000019118 UP000075900 UP000075901 UP000069272 UP000075883 UP000000673 UP000030765 UP000075920 UP000092461 UP000075881 UP000008820 UP000075880 UP000069940 UP000249989 UP000002320 UP000008792 UP000054359
UP000002358 UP000027135 UP000076502 UP000075809 UP000005203 UP000078542 UP000053097 UP000279307 UP000053825 UP000053105 UP000005205 UP000215335 UP000007266 UP000242457 UP000078540 UP000076858 UP000078492 UP000001555 UP000009046 UP000000305 UP000076407 UP000007062 UP000075840 UP000075903 UP000075882 UP000075885 UP000075902 UP000030742 UP000076408 UP000019118 UP000075900 UP000075901 UP000069272 UP000075883 UP000000673 UP000030765 UP000075920 UP000092461 UP000075881 UP000008820 UP000075880 UP000069940 UP000249989 UP000002320 UP000008792 UP000054359
Interpro
IPR013320
ConA-like_dom_sf
+ More
IPR005052 Lectin_leg
IPR035664 VIP36_lectin
IPR027417 P-loop_NTPase
IPR014720 dsRBD_dom
IPR007096 RNA-dir_Rpol_phage_catalytic
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR016137 RGS
IPR005052 Lectin_leg
IPR035664 VIP36_lectin
IPR027417 P-loop_NTPase
IPR014720 dsRBD_dom
IPR007096 RNA-dir_Rpol_phage_catalytic
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR016137 RGS
ProteinModelPortal
H9JMT6
A0A2A4JCQ2
E7EC18
A0A0L7LRV7
A0A2H1WIV8
A0A212EM81
+ More
A0A1E1WTZ7 S4PCC9 A0A194PJU8 E2BH30 K7J8S0 E9ICB0 A0A067R7B1 A0A154PSX6 V9IFA2 A0A151WQX0 A0A088AEZ9 A0A151I8V2 A0A026WBZ9 A0A0L7QKW1 A0A0M8ZUN1 A0A158NXN3 A0A232EYZ5 A0A1B6FSF5 A0A1Y1KZI7 D6X3B0 A0A1B6LHU9 A0A2A3ECC8 A0A1B6IB06 A0A195BRR8 A0A0N8CR05 A0A0P4ZPL0 A0A0P5CR21 J3JZI3 A0A0N7ZPL1 A0A195E4M0 B7QFT9 A0A1B6CZ37 A0A0T6B360 A0A0K8RMH6 E0VUZ8 E9G8W7 A0A293MRX8 A0A0P6AH22 A0A182XF66 Q7QA71 A0A182HVN7 A0A182V8Q8 A0A182KZS1 A0A2R5L9I5 A0A182PLG9 A0A182UAE7 V5IC29 A0A1L8E3B8 U4UM14 A0A182Y909 A0A1L8E362 A0A1L8E2V9 A0A1L8E2X2 N6SYS6 A0A182RI26 A0A182TBZ5 A0A182FAF3 A0A2M3ZKW9 A0A2M4AKU1 A0A2M4BTM6 A0A182MT70 A0A0P5WGH0 T1DNL0 W5JWA7 A0A0P5XHL4 A0A084VDK1 A0A182W383 A0A0P6JRH1 A0A0P5P1W9 A0A1B0GI93 A0A182K4W2 T1ISH8 Q16SI7 A0A224Z396 A0A131Z571 A0A023FYY7 L7LYV5 A0A0P6BXR7 G3MI02 A0A0A9VTR0 A0A182ILX8 A0A023FPA0 A0A182GZZ9 U5ER85 A0A023FER3 T1E2S8 A0A1Z5LBH1 B0W3A9 A0A1Q3F5W4 A0A1Q3F605 B4MBX6 A0A0N8ECK5 A0A0P5TUB7 A0A087TWC9
A0A1E1WTZ7 S4PCC9 A0A194PJU8 E2BH30 K7J8S0 E9ICB0 A0A067R7B1 A0A154PSX6 V9IFA2 A0A151WQX0 A0A088AEZ9 A0A151I8V2 A0A026WBZ9 A0A0L7QKW1 A0A0M8ZUN1 A0A158NXN3 A0A232EYZ5 A0A1B6FSF5 A0A1Y1KZI7 D6X3B0 A0A1B6LHU9 A0A2A3ECC8 A0A1B6IB06 A0A195BRR8 A0A0N8CR05 A0A0P4ZPL0 A0A0P5CR21 J3JZI3 A0A0N7ZPL1 A0A195E4M0 B7QFT9 A0A1B6CZ37 A0A0T6B360 A0A0K8RMH6 E0VUZ8 E9G8W7 A0A293MRX8 A0A0P6AH22 A0A182XF66 Q7QA71 A0A182HVN7 A0A182V8Q8 A0A182KZS1 A0A2R5L9I5 A0A182PLG9 A0A182UAE7 V5IC29 A0A1L8E3B8 U4UM14 A0A182Y909 A0A1L8E362 A0A1L8E2V9 A0A1L8E2X2 N6SYS6 A0A182RI26 A0A182TBZ5 A0A182FAF3 A0A2M3ZKW9 A0A2M4AKU1 A0A2M4BTM6 A0A182MT70 A0A0P5WGH0 T1DNL0 W5JWA7 A0A0P5XHL4 A0A084VDK1 A0A182W383 A0A0P6JRH1 A0A0P5P1W9 A0A1B0GI93 A0A182K4W2 T1ISH8 Q16SI7 A0A224Z396 A0A131Z571 A0A023FYY7 L7LYV5 A0A0P6BXR7 G3MI02 A0A0A9VTR0 A0A182ILX8 A0A023FPA0 A0A182GZZ9 U5ER85 A0A023FER3 T1E2S8 A0A1Z5LBH1 B0W3A9 A0A1Q3F5W4 A0A1Q3F605 B4MBX6 A0A0N8ECK5 A0A0P5TUB7 A0A087TWC9
PDB
2E6V
E-value=4.49338e-76,
Score=723
Ontologies
KEGG
GO
Topology
SignalP
Position: 1 - 30,
Likelihood: 0.744014
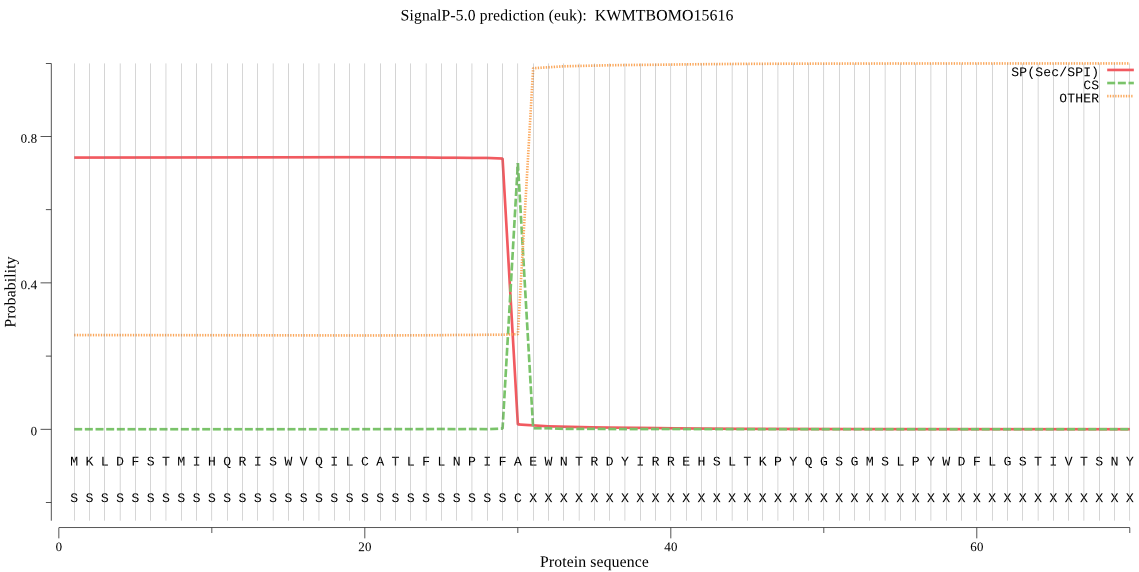
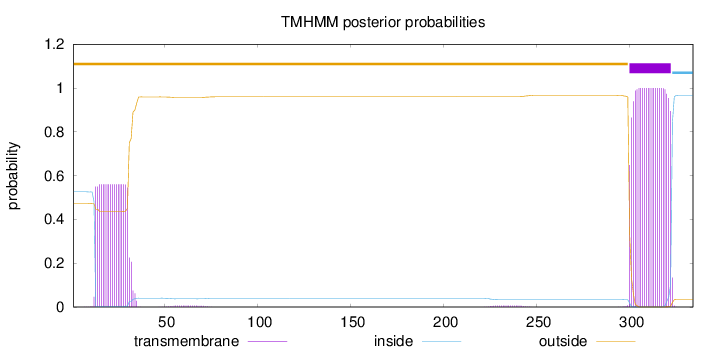
Length:
334
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
33.3552
Exp number, first 60 AAs:
10.75218
Total prob of N-in:
0.52627
POSSIBLE N-term signal
sequence
outside
1 - 299
TMhelix
300 - 322
inside
323 - 334
Population Genetic Test Statistics
Pi
133.753584
Theta
144.556699
Tajima's D
-0.594574
CLR
0.015485
CSRT
0.228188590570471
Interpretation
Uncertain
