Gene
KWMTBOMO15612
Pre Gene Modal
BGIBMGA010717
Annotation
PREDICTED:_osiris_20_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.676
Sequence
CDS
ATGAAACGCGTGTGCATCCTCTTAGCTTCGGTCGCCATCGCGACCTCCTACTCCATTAAGGACAACGAAATCGAACTGCCGAGGCTACGGACGAGTGATGATTTGCTGGACAGTGTTATCAGTGATTGTTCCGAAGCCGGATCGCCCATGGCATGCCTCAAAGTGAAAGTTCTATCATATCTGGACAACAAAGTCGGTGTAGGATCGGAAACGGGGAGGGCATTGGACGAGACAAACATCGACAAAGTCATCTATGATCGGGTGGCGAGGATTTTGGACACCAACGAATTCAAGTTTAAACTGCCGGAGTTTATGTTCCAAAACGCGGAGGTCTCTTACAGAGCCGACAGGGGATTCGATATTGAATTCCCTGAGAACAACGAGAACGCTCGTGGCCTCTTAAAGAAGAAGCTACTGCTTCCCGTCCTGCTGCTCTTGAAGCTTAAAATGAAAGCTATCATGCCGATCCTGGTTTCGATCATCGGTATCAAGGCTATCAAAGCTCTGATCCTCAGTAAGCTGGCCATCACTTTGGTCGTTGGATTCTTGGTTTACAATCTTCTCTTGAAGAAAGGAGGTATGCCAATGATGATGGCTCCGACTGAAGCCCCTCCTGCGCCTCAGTACGGCCCGCCGGCATCCCAATACGGGCCTCCAGCTTCGACCCCAGCAGCGCCCCAGGATTCTTATAGCCCGCAATGGGAGCCTGCAAGCTCTGGACCTTATGCCAGAGTATGGGACCCCTCGCAGCTGGCTTACAGCTCGTACTACCCCGGTGACTCAAGTTCCTCTGCATCAGCGAACCAATCGCCAAGCTACTCGAACGTCGCCTCCATCTCATCATCATCATCGTCATCATCAGCTCCCACCTATTAA
Protein
MKRVCILLASVAIATSYSIKDNEIELPRLRTSDDLLDSVISDCSEAGSPMACLKVKVLSYLDNKVGVGSETGRALDETNIDKVIYDRVARILDTNEFKFKLPEFMFQNAEVSYRADRGFDIEFPENNENARGLLKKKLLLPVLLLLKLKMKAIMPILVSIIGIKAIKALILSKLAITLVVGFLVYNLLLKKGGMPMMMAPTEAPPAPQYGPPASQYGPPASTPAAPQDSYSPQWEPASSGPYARVWDPSQLAYSSYYPGDSSSSASANQSPSYSNVASISSSSSSSSAPTY
Summary
Uniprot
B6DXA7
A0A194PK63
A0A194RQN1
A0A212F7G1
A0A2A4IWP1
A0A0L7L1F4
+ More
A0A2H1VUX5 I4DJL1 A0A1J1HI63 Q298A5 B4G3B6 A0A3B0K2K0 A0A1I8NGW1 A0A240SWM2 A0A1B0B2X4 B4QYA0 B4I4E4 Q9VI07 B4MBN7 B4PVP7 B3NYN8 A0A1A9VDP1 A0A1A9ZSX0 B3LVT4 A0A084WNS4 A0A0K8UXF6 A0A182IX54 B4NHB8 A0A034WS62 B4KBN7 A0A0A1X5W6 W5JJJ5 A0A1W4UQY7 B4JYE0 A0A0M4EMT3 A0A1B0FL94 A0A1A9W5N6 A0A0L0BQB2 Q17GL1 A0A182T3S7 W8C5M4 Q17GL2 A0A1S4F379 A0A1I8JSM1 A0A182G4R4 A0A182YM64 A0A1B0CZP2 A0A182R857 A0A182QQJ7 A0A182MK03 A0A240PNG0 A0A1S4F3F1 A0A1I8MG39 A0A1W4WC97 A0A2P8XGU8 A0A1I8PI59 A0A1Y1LWX8 Q16FK1 A0A1Y9IVG3 A0A067R4N8 J3JYD6 Q7QDF2 A0A1I8NRM6 A0A182XK20 A0A182KX53 A0A182TU75 A0A1I8JTB2 A0A1Y9IVR7 A0A182V8H4 A0A182NKZ5 A0A182KEY1 B0WS66 D6X0X0 A0A2J7PI95 A0A1I8NWF8 A0A336LRU1 E0VX94 A0A232FAW3 K7IU89 A0A154PH50 E2A506 A0A0M9AB34 A0A026WKK3 E2B2X8 A0A3L8DFC6 A0A195C2K0 A0A158NDB5 F4X726 A0A0L7RJG2 A0A151XHH7 A0A088AGH1
A0A2H1VUX5 I4DJL1 A0A1J1HI63 Q298A5 B4G3B6 A0A3B0K2K0 A0A1I8NGW1 A0A240SWM2 A0A1B0B2X4 B4QYA0 B4I4E4 Q9VI07 B4MBN7 B4PVP7 B3NYN8 A0A1A9VDP1 A0A1A9ZSX0 B3LVT4 A0A084WNS4 A0A0K8UXF6 A0A182IX54 B4NHB8 A0A034WS62 B4KBN7 A0A0A1X5W6 W5JJJ5 A0A1W4UQY7 B4JYE0 A0A0M4EMT3 A0A1B0FL94 A0A1A9W5N6 A0A0L0BQB2 Q17GL1 A0A182T3S7 W8C5M4 Q17GL2 A0A1S4F379 A0A1I8JSM1 A0A182G4R4 A0A182YM64 A0A1B0CZP2 A0A182R857 A0A182QQJ7 A0A182MK03 A0A240PNG0 A0A1S4F3F1 A0A1I8MG39 A0A1W4WC97 A0A2P8XGU8 A0A1I8PI59 A0A1Y1LWX8 Q16FK1 A0A1Y9IVG3 A0A067R4N8 J3JYD6 Q7QDF2 A0A1I8NRM6 A0A182XK20 A0A182KX53 A0A182TU75 A0A1I8JTB2 A0A1Y9IVR7 A0A182V8H4 A0A182NKZ5 A0A182KEY1 B0WS66 D6X0X0 A0A2J7PI95 A0A1I8NWF8 A0A336LRU1 E0VX94 A0A232FAW3 K7IU89 A0A154PH50 E2A506 A0A0M9AB34 A0A026WKK3 E2B2X8 A0A3L8DFC6 A0A195C2K0 A0A158NDB5 F4X726 A0A0L7RJG2 A0A151XHH7 A0A088AGH1
Pubmed
19121390
26354079
22118469
26227816
22651552
15632085
+ More
17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588 25348373 25830018 20920257 23761445 26108605 17510324 24495485 26483478 25244985 29403074 28004739 24845553 22516182 12364791 14747013 17210077 20966253 18362917 19820115 20566863 28648823 20075255 20798317 24508170 30249741 21347285 21719571
17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24438588 25348373 25830018 20920257 23761445 26108605 17510324 24495485 26483478 25244985 29403074 28004739 24845553 22516182 12364791 14747013 17210077 20966253 18362917 19820115 20566863 28648823 20075255 20798317 24508170 30249741 21347285 21719571
EMBL
BABH01016839
FJ176295
ACI23617.1
KQ459601
KPI93712.1
KQ459875
+ More
KPJ19620.1 AGBW02009876 OWR49670.1 NWSH01005651 PCG64079.1 JTDY01003617 KOB69255.1 ODYU01004307 SOQ44034.1 AK401479 BAM18101.1 CVRI01000003 CRK87100.1 CM000070 EAL28050.1 CH479179 EDW24298.1 OUUW01000005 SPP80189.1 JXJN01007738 CM000364 EDX11857.1 CH480821 EDW55087.1 AE014297 AY071052 AAF54138.1 AAL48674.1 CH940656 EDW58508.1 CM000160 EDW97856.1 CH954181 EDV48019.1 CH902617 EDV43708.1 ATLV01024676 KE525356 KFB51868.1 GDHF01021053 JAI31261.1 CH964272 EDW84594.1 GAKP01001800 JAC57152.1 CH933806 EDW14714.1 GBXI01007588 JAD06704.1 ADMH02001348 ETN62934.1 CH916377 EDV90702.1 CP012526 ALC46815.1 CCAG010013281 JRES01001623 KNC21389.1 CH477258 EAT45787.1 GAMC01001662 JAC04894.1 EAT45788.1 JXUM01144363 KQ569774 KXJ68519.1 AJVK01020908 AXCN02000107 AXCM01010052 PYGN01002187 PSN31230.1 GEZM01044916 JAV78044.1 CH478422 EAT33017.1 KK852895 KDR14187.1 BT128262 AEE63222.1 AAAB01008851 EAA07389.5 APCN01002211 DS232065 EDS33689.1 KQ971372 EFA10567.1 NEVH01025129 PNF16048.1 UFQT01000139 SSX20726.1 DS235830 EEB18000.1 NNAY01000498 OXU27984.1 AAZX01006235 KQ434902 KZC11127.1 GL436778 EFN71493.1 KQ435706 KOX80186.1 KK107199 EZA55604.1 GL445250 EFN89947.1 QOIP01000009 RLU18892.1 KQ978379 KYM94418.1 ADTU01012542 GL888828 EGI57638.1 KQ414581 KOC70933.1 KQ982130 KYQ59807.1
KPJ19620.1 AGBW02009876 OWR49670.1 NWSH01005651 PCG64079.1 JTDY01003617 KOB69255.1 ODYU01004307 SOQ44034.1 AK401479 BAM18101.1 CVRI01000003 CRK87100.1 CM000070 EAL28050.1 CH479179 EDW24298.1 OUUW01000005 SPP80189.1 JXJN01007738 CM000364 EDX11857.1 CH480821 EDW55087.1 AE014297 AY071052 AAF54138.1 AAL48674.1 CH940656 EDW58508.1 CM000160 EDW97856.1 CH954181 EDV48019.1 CH902617 EDV43708.1 ATLV01024676 KE525356 KFB51868.1 GDHF01021053 JAI31261.1 CH964272 EDW84594.1 GAKP01001800 JAC57152.1 CH933806 EDW14714.1 GBXI01007588 JAD06704.1 ADMH02001348 ETN62934.1 CH916377 EDV90702.1 CP012526 ALC46815.1 CCAG010013281 JRES01001623 KNC21389.1 CH477258 EAT45787.1 GAMC01001662 JAC04894.1 EAT45788.1 JXUM01144363 KQ569774 KXJ68519.1 AJVK01020908 AXCN02000107 AXCM01010052 PYGN01002187 PSN31230.1 GEZM01044916 JAV78044.1 CH478422 EAT33017.1 KK852895 KDR14187.1 BT128262 AEE63222.1 AAAB01008851 EAA07389.5 APCN01002211 DS232065 EDS33689.1 KQ971372 EFA10567.1 NEVH01025129 PNF16048.1 UFQT01000139 SSX20726.1 DS235830 EEB18000.1 NNAY01000498 OXU27984.1 AAZX01006235 KQ434902 KZC11127.1 GL436778 EFN71493.1 KQ435706 KOX80186.1 KK107199 EZA55604.1 GL445250 EFN89947.1 QOIP01000009 RLU18892.1 KQ978379 KYM94418.1 ADTU01012542 GL888828 EGI57638.1 KQ414581 KOC70933.1 KQ982130 KYQ59807.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000037510
+ More
UP000183832 UP000001819 UP000008744 UP000268350 UP000095301 UP000092443 UP000092460 UP000000304 UP000001292 UP000000803 UP000008792 UP000002282 UP000008711 UP000078200 UP000092445 UP000007801 UP000030765 UP000075880 UP000007798 UP000009192 UP000000673 UP000192221 UP000001070 UP000092553 UP000092444 UP000091820 UP000037069 UP000008820 UP000075901 UP000069272 UP000069940 UP000249989 UP000076408 UP000092462 UP000075900 UP000075886 UP000075883 UP000075885 UP000192223 UP000245037 UP000095300 UP000075920 UP000027135 UP000007062 UP000076407 UP000075882 UP000075902 UP000075840 UP000075903 UP000075884 UP000075881 UP000002320 UP000007266 UP000235965 UP000009046 UP000215335 UP000002358 UP000076502 UP000000311 UP000053105 UP000053097 UP000008237 UP000279307 UP000078542 UP000005205 UP000007755 UP000053825 UP000075809 UP000005203
UP000183832 UP000001819 UP000008744 UP000268350 UP000095301 UP000092443 UP000092460 UP000000304 UP000001292 UP000000803 UP000008792 UP000002282 UP000008711 UP000078200 UP000092445 UP000007801 UP000030765 UP000075880 UP000007798 UP000009192 UP000000673 UP000192221 UP000001070 UP000092553 UP000092444 UP000091820 UP000037069 UP000008820 UP000075901 UP000069272 UP000069940 UP000249989 UP000076408 UP000092462 UP000075900 UP000075886 UP000075883 UP000075885 UP000192223 UP000245037 UP000095300 UP000075920 UP000027135 UP000007062 UP000076407 UP000075882 UP000075902 UP000075840 UP000075903 UP000075884 UP000075881 UP000002320 UP000007266 UP000235965 UP000009046 UP000215335 UP000002358 UP000076502 UP000000311 UP000053105 UP000053097 UP000008237 UP000279307 UP000078542 UP000005205 UP000007755 UP000053825 UP000075809 UP000005203
Interpro
SUPFAM
SSF49777
SSF49777
Gene 3D
CDD
ProteinModelPortal
B6DXA7
A0A194PK63
A0A194RQN1
A0A212F7G1
A0A2A4IWP1
A0A0L7L1F4
+ More
A0A2H1VUX5 I4DJL1 A0A1J1HI63 Q298A5 B4G3B6 A0A3B0K2K0 A0A1I8NGW1 A0A240SWM2 A0A1B0B2X4 B4QYA0 B4I4E4 Q9VI07 B4MBN7 B4PVP7 B3NYN8 A0A1A9VDP1 A0A1A9ZSX0 B3LVT4 A0A084WNS4 A0A0K8UXF6 A0A182IX54 B4NHB8 A0A034WS62 B4KBN7 A0A0A1X5W6 W5JJJ5 A0A1W4UQY7 B4JYE0 A0A0M4EMT3 A0A1B0FL94 A0A1A9W5N6 A0A0L0BQB2 Q17GL1 A0A182T3S7 W8C5M4 Q17GL2 A0A1S4F379 A0A1I8JSM1 A0A182G4R4 A0A182YM64 A0A1B0CZP2 A0A182R857 A0A182QQJ7 A0A182MK03 A0A240PNG0 A0A1S4F3F1 A0A1I8MG39 A0A1W4WC97 A0A2P8XGU8 A0A1I8PI59 A0A1Y1LWX8 Q16FK1 A0A1Y9IVG3 A0A067R4N8 J3JYD6 Q7QDF2 A0A1I8NRM6 A0A182XK20 A0A182KX53 A0A182TU75 A0A1I8JTB2 A0A1Y9IVR7 A0A182V8H4 A0A182NKZ5 A0A182KEY1 B0WS66 D6X0X0 A0A2J7PI95 A0A1I8NWF8 A0A336LRU1 E0VX94 A0A232FAW3 K7IU89 A0A154PH50 E2A506 A0A0M9AB34 A0A026WKK3 E2B2X8 A0A3L8DFC6 A0A195C2K0 A0A158NDB5 F4X726 A0A0L7RJG2 A0A151XHH7 A0A088AGH1
A0A2H1VUX5 I4DJL1 A0A1J1HI63 Q298A5 B4G3B6 A0A3B0K2K0 A0A1I8NGW1 A0A240SWM2 A0A1B0B2X4 B4QYA0 B4I4E4 Q9VI07 B4MBN7 B4PVP7 B3NYN8 A0A1A9VDP1 A0A1A9ZSX0 B3LVT4 A0A084WNS4 A0A0K8UXF6 A0A182IX54 B4NHB8 A0A034WS62 B4KBN7 A0A0A1X5W6 W5JJJ5 A0A1W4UQY7 B4JYE0 A0A0M4EMT3 A0A1B0FL94 A0A1A9W5N6 A0A0L0BQB2 Q17GL1 A0A182T3S7 W8C5M4 Q17GL2 A0A1S4F379 A0A1I8JSM1 A0A182G4R4 A0A182YM64 A0A1B0CZP2 A0A182R857 A0A182QQJ7 A0A182MK03 A0A240PNG0 A0A1S4F3F1 A0A1I8MG39 A0A1W4WC97 A0A2P8XGU8 A0A1I8PI59 A0A1Y1LWX8 Q16FK1 A0A1Y9IVG3 A0A067R4N8 J3JYD6 Q7QDF2 A0A1I8NRM6 A0A182XK20 A0A182KX53 A0A182TU75 A0A1I8JTB2 A0A1Y9IVR7 A0A182V8H4 A0A182NKZ5 A0A182KEY1 B0WS66 D6X0X0 A0A2J7PI95 A0A1I8NWF8 A0A336LRU1 E0VX94 A0A232FAW3 K7IU89 A0A154PH50 E2A506 A0A0M9AB34 A0A026WKK3 E2B2X8 A0A3L8DFC6 A0A195C2K0 A0A158NDB5 F4X726 A0A0L7RJG2 A0A151XHH7 A0A088AGH1
Ontologies
PANTHER
Topology
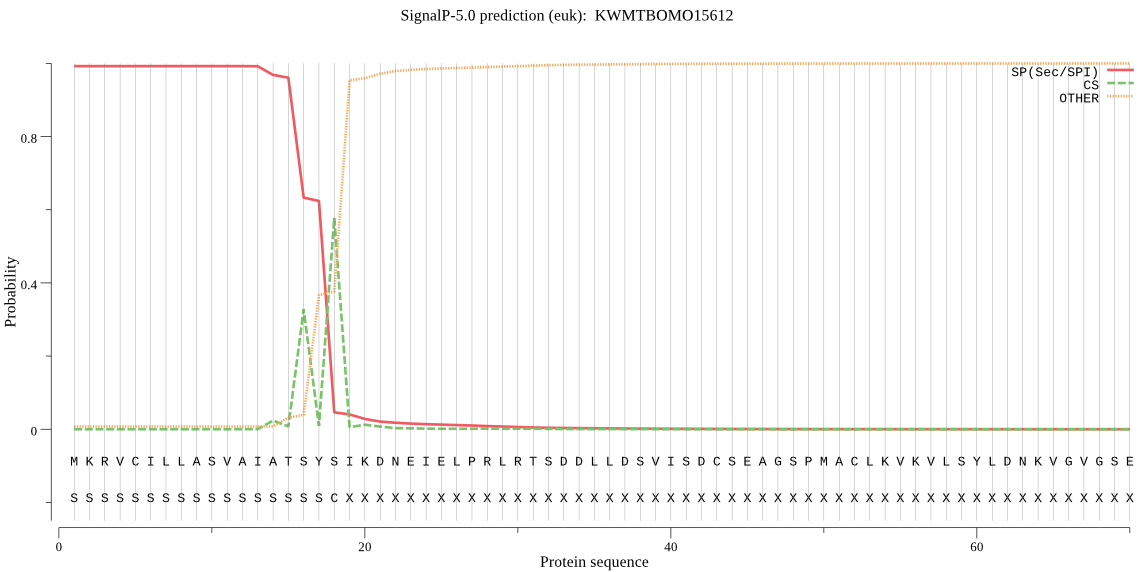
SignalP
Position: 1 - 18,
Likelihood: 0.992291
Length:
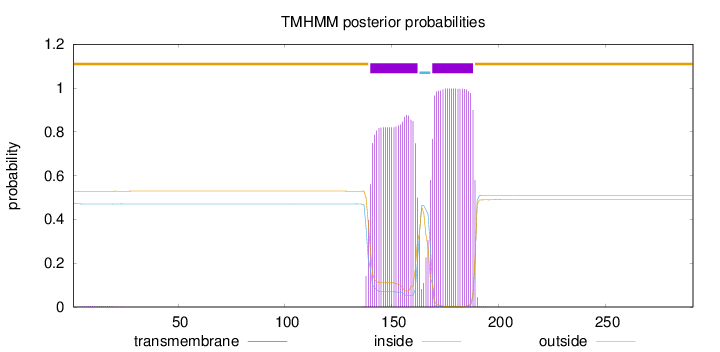
291
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.72125
Exp number, first 60 AAs:
0.04509
Total prob of N-in:
0.47192
outside
1 - 139
TMhelix
140 - 162
inside
163 - 168
TMhelix
169 - 188
outside
189 - 291
Population Genetic Test Statistics
Pi
141.878999
Theta
133.029836
Tajima's D
0.246268
CLR
0.304367
CSRT
0.43762811859407
Interpretation
Uncertain
