Gene
KWMTBOMO15610
Pre Gene Modal
BGIBMGA010837
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_equilibrative_nucleoside_transporter_1_[Papilio_machaon]
Full name
NADPH-dependent diflavin oxidoreductase 1
Alternative Name
NADPH-dependent FMN and FAD-containing oxidoreductase
Location in the cell
PlasmaMembrane Reliability : 2.292
Sequence
CDS
ATGGAGATGAGCCATCAAGGTGGGGCGGAGGCCGAGAGCCTCCTAAGACATGACCAGAGGAGCGCCATCGTCATGCCCGGCCAGCGGACTGGACGCTGGGAGGATGGCGAGAAGGCTCCGTTCCTACGTAACGAGCCGGTGAAACTGACACCCGCTTGGGAGGCCACGAACCTGCCCAATGACACGCTCAACTTGAAAGGTATAGCAATGGACCTGACTCCGCCGAAAGATAAATGGAACCTCATTTATCTGACGTTGGTGCTTCACGGCTTGGGAACGCTGACCGCCTGGAACATGTTCATAACAGCGAAAGCGTACTTTGAGACGTACAAACTCGCCTCGGAGCCGGCCTTGTCCGAGAACTTCCTCACGTACATCGGCTGGGCGAGTCAGATACCGAATTTGCTGTTCAGCTGGTTTAATATATTTTTCCAATTGGGGTATGTTAATTTTTTTTTATTTTTTATTGTTTAG
Protein
MEMSHQGGAEAESLLRHDQRSAIVMPGQRTGRWEDGEKAPFLRNEPVKLTPAWEATNLPNDTLNLKGIAMDLTPPKDKWNLIYLTLVLHGLGTLTAWNMFITAKAYFETYKLASEPALSENFLTYIGWASQIPNLLFSWFNIFFQLGYVNFFLFFIV
Summary
Description
Component of the cytosolic iron-sulfur (Fe-S) protein assembly (CIA) machinery. Required for the maturation of extramitochondrial Fe-S proteins. Part of an electron transfer chain functioning in an early step of cytosolic Fe-S biogenesis. Transfers electrons from NADPH to the Fe-S cluster of the anamorsin/DRE2 homolog.
Cofactor
FAD
FMN
FMN
Similarity
Belongs to the NADPH-dependent diflavin oxidoreductase NDOR1 family.
In the C-terminal section; belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
In the N-terminal section; belongs to the flavodoxin family.
In the C-terminal section; belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
In the N-terminal section; belongs to the flavodoxin family.
Uniprot
A0A194PL39
H9JMT4
S4PKB3
A0A212EYJ3
A0A3S2NQK3
A0A194RQE1
+ More
A0A023EU60 A0A1S4FAG4 A0A336MBT2 A0A1Q3FFC7 A0A1Q3FFH8 A0A1B6JU47 A0A1B6FRU4 A0A1B6DIQ1 A0A336MEK9 A0A1B6LYW3 A0A182VQC5 B0WGR4 A0A1L8DEK0 A0A1L8DEI1 A0A1L8DEI3 A0A182LWW1 A0A1B6M312 A0A1B0CMU0 A0A023F3B9 A0A224XFF7 A0A0K8TKS5 A0A1B0AKG2 A0A182IXK0 A0A182N6W6 A0A1A9VM26 A0A164I0D6 A0A182RBV4 A0A146MH54 A0A0A9WT49 A0A0K8SUD5 A0A3Q0J2I2 A0A195F335 A0A182TDX6 Q7PTI1 A0A0J7L8Y2 A0A182K078 A0A182LAE8 A0A182XX87 U5EVT7 A0A2R7VPU9 A0A182Q2X6 A0A1I8PKL2 E2BV50 A0A0P6H3N9 A0A0P4YWH3 A0A1J1HM94 A0A151J0K3 A0A0P5A9T3 T1HNN9 A0A0P6BJ91 T1EAP4 A0A2M4CHE9 A0A0M9A978 A0A182F8G7 V9I984 A0A2M4BKX5 A0A0L7R2K6 T1PI77 A0A2M4BL33 A0A154PJX0 A0A158NZY4 A0A2A3E229 A0A087ZWM1 A0A195CNP2 A0A151X4G1 A0A026WZJ9 A0A182PBD6 E9FRB5 A0A3L8DRB1 F4WII4 A0A2J7PI52 A0A2J7PI60 E9J590 A0A310SI93 A0A084WQY1 K7INK5 A0A182I8J1 A0A182WYG2 A0A067R482 A0A232EU38 A0A1Y1LTG1 J9K041 D6X319 A0A2S2QTU9 A0A0C9R913 A0A2T7PBP1 A0A0T6AWR2 A0A1B6FGI1 E0W0A7
A0A023EU60 A0A1S4FAG4 A0A336MBT2 A0A1Q3FFC7 A0A1Q3FFH8 A0A1B6JU47 A0A1B6FRU4 A0A1B6DIQ1 A0A336MEK9 A0A1B6LYW3 A0A182VQC5 B0WGR4 A0A1L8DEK0 A0A1L8DEI1 A0A1L8DEI3 A0A182LWW1 A0A1B6M312 A0A1B0CMU0 A0A023F3B9 A0A224XFF7 A0A0K8TKS5 A0A1B0AKG2 A0A182IXK0 A0A182N6W6 A0A1A9VM26 A0A164I0D6 A0A182RBV4 A0A146MH54 A0A0A9WT49 A0A0K8SUD5 A0A3Q0J2I2 A0A195F335 A0A182TDX6 Q7PTI1 A0A0J7L8Y2 A0A182K078 A0A182LAE8 A0A182XX87 U5EVT7 A0A2R7VPU9 A0A182Q2X6 A0A1I8PKL2 E2BV50 A0A0P6H3N9 A0A0P4YWH3 A0A1J1HM94 A0A151J0K3 A0A0P5A9T3 T1HNN9 A0A0P6BJ91 T1EAP4 A0A2M4CHE9 A0A0M9A978 A0A182F8G7 V9I984 A0A2M4BKX5 A0A0L7R2K6 T1PI77 A0A2M4BL33 A0A154PJX0 A0A158NZY4 A0A2A3E229 A0A087ZWM1 A0A195CNP2 A0A151X4G1 A0A026WZJ9 A0A182PBD6 E9FRB5 A0A3L8DRB1 F4WII4 A0A2J7PI52 A0A2J7PI60 E9J590 A0A310SI93 A0A084WQY1 K7INK5 A0A182I8J1 A0A182WYG2 A0A067R482 A0A232EU38 A0A1Y1LTG1 J9K041 D6X319 A0A2S2QTU9 A0A0C9R913 A0A2T7PBP1 A0A0T6AWR2 A0A1B6FGI1 E0W0A7
EC Number
1.18.1.-
Pubmed
EMBL
KQ459601
KPI93713.1
BABH01016835
BABH01016836
BABH01016837
GAIX01004595
+ More
JAA87965.1 AGBW02011486 OWR46568.1 RSAL01000010 RVE53678.1 KQ459875 KPJ19619.1 GAPW01001136 JAC12462.1 UFQT01000883 SSX27822.1 GFDL01008802 JAV26243.1 GFDL01008783 JAV26262.1 GECU01005024 JAT02683.1 GECZ01016861 JAS52908.1 GEDC01011732 JAS25566.1 SSX27821.1 GEBQ01011101 JAT28876.1 DS231928 EDS27140.1 GFDF01009213 JAV04871.1 GFDF01009211 JAV04873.1 GFDF01009212 JAV04872.1 AXCM01004381 GEBQ01009661 JAT30316.1 AJWK01019162 AJWK01019163 AJWK01019164 AJWK01019165 GBBI01002939 JAC15773.1 GFTR01006662 JAW09764.1 GDAI01002679 JAI14924.1 JXJN01025262 LRGB01008684 KZS00747.1 GDHC01020303 GDHC01000352 JAP98325.1 JAQ18277.1 GBHO01035571 GBHO01035569 GBHO01035568 GBHO01035567 JAG08033.1 JAG08035.1 JAG08036.1 JAG08037.1 GBRD01008898 JAG56923.1 KQ981856 KYN34592.1 AAAB01008805 EAA03890.5 LBMM01000214 KMR04460.1 GANO01000868 JAB59003.1 KK854007 PTY09168.1 AXCN02000310 GL450810 EFN80440.1 GDIQ01024555 JAN70182.1 GDIP01222640 JAJ00762.1 CVRI01000008 CRK88524.1 KQ980624 KYN15035.1 GDIP01202133 JAJ21269.1 ACPB03007647 GDIP01015037 JAM88678.1 GAMD01000232 JAB01359.1 GGFL01000555 MBW64733.1 KQ435721 KOX78523.1 JR037241 AEY57658.1 GGFJ01004564 MBW53705.1 KQ414666 KOC65069.1 KA647618 AFP62247.1 GGFJ01004563 MBW53704.1 KQ434938 KZC12156.1 ADTU01005131 ADTU01005132 KZ288438 PBC25745.1 KQ977580 KYN01719.1 KQ982557 KYQ55108.1 KK107063 EZA61258.1 GL732523 EFX90137.1 QOIP01000005 RLU22974.1 GL888175 EGI66115.1 NEVH01025129 PNF16026.1 PNF16025.1 GL768124 EFZ12035.1 KQ768431 OAD53172.1 ATLV01025806 ATLV01025807 ATLV01025808 KE525400 KFB52625.1 APCN01003039 KK852766 KDR16974.1 NNAY01002170 OXU21879.1 GEZM01050427 JAV75590.1 ABLF02024980 KQ971372 EFA10308.1 GGMS01011951 MBY81154.1 GBYB01009342 JAG79109.1 PZQS01000005 PVD30835.1 LJIG01022679 KRT79285.1 GECZ01020457 JAS49312.1 DS235858 EEB19063.1
JAA87965.1 AGBW02011486 OWR46568.1 RSAL01000010 RVE53678.1 KQ459875 KPJ19619.1 GAPW01001136 JAC12462.1 UFQT01000883 SSX27822.1 GFDL01008802 JAV26243.1 GFDL01008783 JAV26262.1 GECU01005024 JAT02683.1 GECZ01016861 JAS52908.1 GEDC01011732 JAS25566.1 SSX27821.1 GEBQ01011101 JAT28876.1 DS231928 EDS27140.1 GFDF01009213 JAV04871.1 GFDF01009211 JAV04873.1 GFDF01009212 JAV04872.1 AXCM01004381 GEBQ01009661 JAT30316.1 AJWK01019162 AJWK01019163 AJWK01019164 AJWK01019165 GBBI01002939 JAC15773.1 GFTR01006662 JAW09764.1 GDAI01002679 JAI14924.1 JXJN01025262 LRGB01008684 KZS00747.1 GDHC01020303 GDHC01000352 JAP98325.1 JAQ18277.1 GBHO01035571 GBHO01035569 GBHO01035568 GBHO01035567 JAG08033.1 JAG08035.1 JAG08036.1 JAG08037.1 GBRD01008898 JAG56923.1 KQ981856 KYN34592.1 AAAB01008805 EAA03890.5 LBMM01000214 KMR04460.1 GANO01000868 JAB59003.1 KK854007 PTY09168.1 AXCN02000310 GL450810 EFN80440.1 GDIQ01024555 JAN70182.1 GDIP01222640 JAJ00762.1 CVRI01000008 CRK88524.1 KQ980624 KYN15035.1 GDIP01202133 JAJ21269.1 ACPB03007647 GDIP01015037 JAM88678.1 GAMD01000232 JAB01359.1 GGFL01000555 MBW64733.1 KQ435721 KOX78523.1 JR037241 AEY57658.1 GGFJ01004564 MBW53705.1 KQ414666 KOC65069.1 KA647618 AFP62247.1 GGFJ01004563 MBW53704.1 KQ434938 KZC12156.1 ADTU01005131 ADTU01005132 KZ288438 PBC25745.1 KQ977580 KYN01719.1 KQ982557 KYQ55108.1 KK107063 EZA61258.1 GL732523 EFX90137.1 QOIP01000005 RLU22974.1 GL888175 EGI66115.1 NEVH01025129 PNF16026.1 PNF16025.1 GL768124 EFZ12035.1 KQ768431 OAD53172.1 ATLV01025806 ATLV01025807 ATLV01025808 KE525400 KFB52625.1 APCN01003039 KK852766 KDR16974.1 NNAY01002170 OXU21879.1 GEZM01050427 JAV75590.1 ABLF02024980 KQ971372 EFA10308.1 GGMS01011951 MBY81154.1 GBYB01009342 JAG79109.1 PZQS01000005 PVD30835.1 LJIG01022679 KRT79285.1 GECZ01020457 JAS49312.1 DS235858 EEB19063.1
Proteomes
UP000053268
UP000005204
UP000007151
UP000283053
UP000053240
UP000075920
+ More
UP000002320 UP000075883 UP000092461 UP000092460 UP000075880 UP000075884 UP000078200 UP000076858 UP000075900 UP000079169 UP000078541 UP000075902 UP000007062 UP000036403 UP000075881 UP000075882 UP000076408 UP000075886 UP000095300 UP000008237 UP000183832 UP000078492 UP000015103 UP000053105 UP000069272 UP000053825 UP000095301 UP000076502 UP000005205 UP000242457 UP000005203 UP000078542 UP000075809 UP000053097 UP000075885 UP000000305 UP000279307 UP000007755 UP000235965 UP000030765 UP000002358 UP000075840 UP000076407 UP000027135 UP000215335 UP000007819 UP000007266 UP000245119 UP000009046
UP000002320 UP000075883 UP000092461 UP000092460 UP000075880 UP000075884 UP000078200 UP000076858 UP000075900 UP000079169 UP000078541 UP000075902 UP000007062 UP000036403 UP000075881 UP000075882 UP000076408 UP000075886 UP000095300 UP000008237 UP000183832 UP000078492 UP000015103 UP000053105 UP000069272 UP000053825 UP000095301 UP000076502 UP000005205 UP000242457 UP000005203 UP000078542 UP000075809 UP000053097 UP000075885 UP000000305 UP000279307 UP000007755 UP000235965 UP000030765 UP000002358 UP000075840 UP000076407 UP000027135 UP000215335 UP000007819 UP000007266 UP000245119 UP000009046
PRIDE
Pfam
Interpro
IPR002259
Eqnu_transpt
+ More
IPR036259 MFS_trans_sf
IPR000300 IPPc
IPR002013 SAC_dom
IPR015047 DUF1866
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR034972 SYNJ1
IPR020846 MFS_dom
IPR023173 NADPH_Cyt_P450_Rdtase_alpha
IPR028879 NDOR1
IPR039261 FNR_nucleotide-bd
IPR003097 CysJ-like_FAD-binding
IPR017938 Riboflavin_synthase-like_b-brl
IPR008254 Flavodoxin/NO_synth
IPR001433 OxRdtase_FAD/NAD-bd
IPR017927 FAD-bd_FR_type
IPR029039 Flavoprotein-like_sf
IPR001094 Flavdoxin-like
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
IPR036259 MFS_trans_sf
IPR000300 IPPc
IPR002013 SAC_dom
IPR015047 DUF1866
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR034972 SYNJ1
IPR020846 MFS_dom
IPR023173 NADPH_Cyt_P450_Rdtase_alpha
IPR028879 NDOR1
IPR039261 FNR_nucleotide-bd
IPR003097 CysJ-like_FAD-binding
IPR017938 Riboflavin_synthase-like_b-brl
IPR008254 Flavodoxin/NO_synth
IPR001433 OxRdtase_FAD/NAD-bd
IPR017927 FAD-bd_FR_type
IPR029039 Flavoprotein-like_sf
IPR001094 Flavdoxin-like
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A194PL39
H9JMT4
S4PKB3
A0A212EYJ3
A0A3S2NQK3
A0A194RQE1
+ More
A0A023EU60 A0A1S4FAG4 A0A336MBT2 A0A1Q3FFC7 A0A1Q3FFH8 A0A1B6JU47 A0A1B6FRU4 A0A1B6DIQ1 A0A336MEK9 A0A1B6LYW3 A0A182VQC5 B0WGR4 A0A1L8DEK0 A0A1L8DEI1 A0A1L8DEI3 A0A182LWW1 A0A1B6M312 A0A1B0CMU0 A0A023F3B9 A0A224XFF7 A0A0K8TKS5 A0A1B0AKG2 A0A182IXK0 A0A182N6W6 A0A1A9VM26 A0A164I0D6 A0A182RBV4 A0A146MH54 A0A0A9WT49 A0A0K8SUD5 A0A3Q0J2I2 A0A195F335 A0A182TDX6 Q7PTI1 A0A0J7L8Y2 A0A182K078 A0A182LAE8 A0A182XX87 U5EVT7 A0A2R7VPU9 A0A182Q2X6 A0A1I8PKL2 E2BV50 A0A0P6H3N9 A0A0P4YWH3 A0A1J1HM94 A0A151J0K3 A0A0P5A9T3 T1HNN9 A0A0P6BJ91 T1EAP4 A0A2M4CHE9 A0A0M9A978 A0A182F8G7 V9I984 A0A2M4BKX5 A0A0L7R2K6 T1PI77 A0A2M4BL33 A0A154PJX0 A0A158NZY4 A0A2A3E229 A0A087ZWM1 A0A195CNP2 A0A151X4G1 A0A026WZJ9 A0A182PBD6 E9FRB5 A0A3L8DRB1 F4WII4 A0A2J7PI52 A0A2J7PI60 E9J590 A0A310SI93 A0A084WQY1 K7INK5 A0A182I8J1 A0A182WYG2 A0A067R482 A0A232EU38 A0A1Y1LTG1 J9K041 D6X319 A0A2S2QTU9 A0A0C9R913 A0A2T7PBP1 A0A0T6AWR2 A0A1B6FGI1 E0W0A7
A0A023EU60 A0A1S4FAG4 A0A336MBT2 A0A1Q3FFC7 A0A1Q3FFH8 A0A1B6JU47 A0A1B6FRU4 A0A1B6DIQ1 A0A336MEK9 A0A1B6LYW3 A0A182VQC5 B0WGR4 A0A1L8DEK0 A0A1L8DEI1 A0A1L8DEI3 A0A182LWW1 A0A1B6M312 A0A1B0CMU0 A0A023F3B9 A0A224XFF7 A0A0K8TKS5 A0A1B0AKG2 A0A182IXK0 A0A182N6W6 A0A1A9VM26 A0A164I0D6 A0A182RBV4 A0A146MH54 A0A0A9WT49 A0A0K8SUD5 A0A3Q0J2I2 A0A195F335 A0A182TDX6 Q7PTI1 A0A0J7L8Y2 A0A182K078 A0A182LAE8 A0A182XX87 U5EVT7 A0A2R7VPU9 A0A182Q2X6 A0A1I8PKL2 E2BV50 A0A0P6H3N9 A0A0P4YWH3 A0A1J1HM94 A0A151J0K3 A0A0P5A9T3 T1HNN9 A0A0P6BJ91 T1EAP4 A0A2M4CHE9 A0A0M9A978 A0A182F8G7 V9I984 A0A2M4BKX5 A0A0L7R2K6 T1PI77 A0A2M4BL33 A0A154PJX0 A0A158NZY4 A0A2A3E229 A0A087ZWM1 A0A195CNP2 A0A151X4G1 A0A026WZJ9 A0A182PBD6 E9FRB5 A0A3L8DRB1 F4WII4 A0A2J7PI52 A0A2J7PI60 E9J590 A0A310SI93 A0A084WQY1 K7INK5 A0A182I8J1 A0A182WYG2 A0A067R482 A0A232EU38 A0A1Y1LTG1 J9K041 D6X319 A0A2S2QTU9 A0A0C9R913 A0A2T7PBP1 A0A0T6AWR2 A0A1B6FGI1 E0W0A7
Ontologies
GO
PANTHER
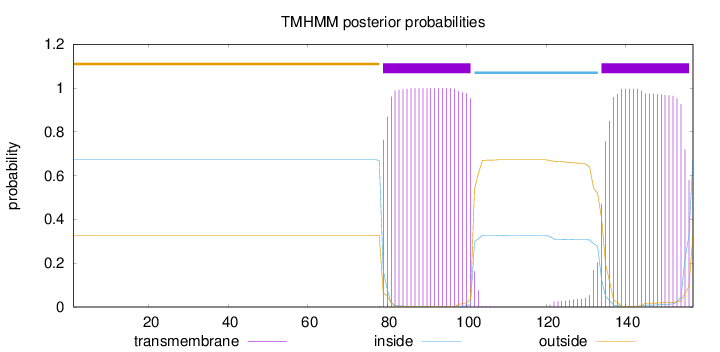
Topology
Subcellular location
Cytoplasm
Length:
157
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.34927
Exp number, first 60 AAs:
0
Total prob of N-in:
0.67293
outside
1 - 78
TMhelix
79 - 101
inside
102 - 133
TMhelix
134 - 156
outside
157 - 157
Population Genetic Test Statistics
Pi
212.974639
Theta
166.284062
Tajima's D
0.915673
CLR
0.097905
CSRT
0.632018399080046
Interpretation
Uncertain
