Gene
KWMTBOMO15608
Annotation
PREDICTED:_zinc_finger_protein_37A-like_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.415
Sequence
CDS
ATGCCGCTTGTTGCATATAAGGAGAAAAAAAAACTAAGAATCCAATGTAATCTCTGTAAAGCGATAATCAGATCTGACTCATACCAAAAACATTTGATGCGGCATCAGTCAGCAACTCACTTCGTATGTGAAGTGTGTGGCAAACTATATCGGAAAGACAACCTGATCAGACACTTACAGTTACACAGTGATTATTTACCACATGTGTGCCAAATATGTCCATACCGAGGCAGATTCTATGAGTCATTAAAAATACATTTACGTACCCACAGTGGTGACAAGCCTTTTTCCTGTGACAAGTGTAGTCTAAGATTTTTGACACGAAGCAACCTCAACAGGCATCTTCTGACTCATAAAAAAGAAAAACCCTTTAAATGCTTGGAATGTAGTCGAAGTTTCTATTTGAAGAGAGACATGGAAGCACATTTTAAAGCAGATCATACAGGTGTAAAGGATTTTGTATGTGATGCCTGCGGCAACAAGTATGGAACAAAGAAGGCATTGATGAGACATGAACTTAGAGTACACAAAAGAGACAAAATGACAAAGGGGCGAACACCTCTGTACTTATTAAATAACAGAAAATGCAATGGAGTCCAATAG
Protein
MPLVAYKEKKKLRIQCNLCKAIIRSDSYQKHLMRHQSATHFVCEVCGKLYRKDNLIRHLQLHSDYLPHVCQICPYRGRFYESLKIHLRTHSGDKPFSCDKCSLRFLTRSNLNRHLLTHKKEKPFKCLECSRSFYLKRDMEAHFKADHTGVKDFVCDACGNKYGTKKALMRHELRVHKRDKMTKGRTPLYLLNNRKCNGVQ
Summary
Uniprot
A0A2H1VT86
A0A2A4JG74
A0A1E1WIZ6
A0A212F2U4
H9JMG1
A0A0L7LRQ9
+ More
A0A2H1W341 S4PGP2 A0A194PK55 A0A194RP61 A0A2A4JFN6 A0A194RQM6 A0A194PJV7 A0A1E1W6T3 A0A2A4JER5 A0A0L7KN53 A0A212EYL8 A0A1E1WPA8 H9JMG6 A0A2H1WH54 A0A1E1WJI4 S4PD46 A0A1E1W1P5 A0A2A4JEU2 H9JMG5 A0A0L7K2S7 A0A3Q0SQ85 A0A3Q2R090 A0A146VRK5 A0A0L7KNS3 A0A146ZUX2 A0A146PAU9 A0A146PMY2 A0A2R8Q0I5 A0A3Q3VV05 A0A3N0YIY5 A0A3B3XSK4 A0A212FG77 B3NX02 M9PHD3 Q9W3J2 A0A2I4BKY4 A0A3Q3AJH2 B4Q0F1 A0A2I4AJI5 A0A1B6I224 A0A3B5KUY2 A0A3Q3FVC2 A0A3B4Y2T3 A0A1A8DZZ5 A0A0S7JXW1 A0A147AE88 A0A2R8QH71 A0A3B3TK93 A0A3B5QTA6 A0A2I4BIT0 A0A1A8DJN5 A0A096M862 A0A096M2C9 A0A3Q3R3T5 S4NVL5 A0A3Q2PXS2 A0A146SJ19 A0A3Q3DHP3 A0A3Q3F3C3 A0A2P8YWR9 A0A087X5E0 A0A146XBG6 A0A3Q2UIJ1 A0A147ALR8 A0A3B3YA57 A0A3Q3A8R5 A0A146ZJI3 A0A146XDR0 A0A146SBU3 A0A146RF51 A0A087XBF4 A0A3Q4GBK2 A0A0G2KSV7 V4ANB0 A0A146QR30 A0A146Q7L4 A0A315W029 A0A1W4VWH2 A0A3Q2PS42 A0A3B5PVQ2 A0A146QK88 B4R6G4 A0A3Q2XZ96 A0A3B3UY54 B4IKX7 A0A3P9BXK5 H9J326 A0A3B3UEH5 A0A1A7ZVQ2 A0A146SEV3 A0A3B4H1W0 A0A146PUD5 A0A3Q3CRW7
A0A2H1W341 S4PGP2 A0A194PK55 A0A194RP61 A0A2A4JFN6 A0A194RQM6 A0A194PJV7 A0A1E1W6T3 A0A2A4JER5 A0A0L7KN53 A0A212EYL8 A0A1E1WPA8 H9JMG6 A0A2H1WH54 A0A1E1WJI4 S4PD46 A0A1E1W1P5 A0A2A4JEU2 H9JMG5 A0A0L7K2S7 A0A3Q0SQ85 A0A3Q2R090 A0A146VRK5 A0A0L7KNS3 A0A146ZUX2 A0A146PAU9 A0A146PMY2 A0A2R8Q0I5 A0A3Q3VV05 A0A3N0YIY5 A0A3B3XSK4 A0A212FG77 B3NX02 M9PHD3 Q9W3J2 A0A2I4BKY4 A0A3Q3AJH2 B4Q0F1 A0A2I4AJI5 A0A1B6I224 A0A3B5KUY2 A0A3Q3FVC2 A0A3B4Y2T3 A0A1A8DZZ5 A0A0S7JXW1 A0A147AE88 A0A2R8QH71 A0A3B3TK93 A0A3B5QTA6 A0A2I4BIT0 A0A1A8DJN5 A0A096M862 A0A096M2C9 A0A3Q3R3T5 S4NVL5 A0A3Q2PXS2 A0A146SJ19 A0A3Q3DHP3 A0A3Q3F3C3 A0A2P8YWR9 A0A087X5E0 A0A146XBG6 A0A3Q2UIJ1 A0A147ALR8 A0A3B3YA57 A0A3Q3A8R5 A0A146ZJI3 A0A146XDR0 A0A146SBU3 A0A146RF51 A0A087XBF4 A0A3Q4GBK2 A0A0G2KSV7 V4ANB0 A0A146QR30 A0A146Q7L4 A0A315W029 A0A1W4VWH2 A0A3Q2PS42 A0A3B5PVQ2 A0A146QK88 B4R6G4 A0A3Q2XZ96 A0A3B3UY54 B4IKX7 A0A3P9BXK5 H9J326 A0A3B3UEH5 A0A1A7ZVQ2 A0A146SEV3 A0A3B4H1W0 A0A146PUD5 A0A3Q3CRW7
Pubmed
EMBL
ODYU01004307
SOQ44037.1
NWSH01001646
PCG70574.1
GDQN01004061
JAT86993.1
+ More
AGBW02010685 OWR48043.1 BABH01016863 JTDY01000233 KOB78143.1 ODYU01005946 SOQ47366.1 GAIX01003572 JAA88988.1 KQ459601 KPI93702.1 KQ459875 KPJ19633.1 PCG70576.1 KPJ19615.1 KPI93716.1 GDQN01008348 JAT82706.1 PCG70575.1 JTDY01008160 KOB64707.1 AGBW02011486 OWR46570.1 GDQN01002209 JAT88845.1 BABH01016826 ODYU01008528 SOQ52212.1 GDQN01003874 JAT87180.1 GAIX01003806 JAA88754.1 GDQN01010134 JAT80920.1 PCG70577.1 BABH01016827 JTDY01015286 KOB51959.1 GCES01066756 JAR19567.1 KOB64706.1 GCES01016586 JAR69737.1 GCES01145374 JAQ40948.1 GCES01140993 JAQ45329.1 CR790382 RJVU01040460 ROL46202.1 AGBW02008703 OWR52736.1 CH954180 EDV46831.1 AE014298 AGB95169.1 AY061144 AAF46335.1 AAL28692.1 CM000162 EDX02288.1 GECU01026801 JAS80905.1 HAEA01011435 SBQ39915.1 GBYX01226456 JAO70000.1 GCES01009514 JAR76809.1 HAEA01006085 SBQ34565.1 AYCK01020751 AYCK01020752 AYCK01020753 AYCK01020754 AYCK01019130 GAIX01009769 JAA82791.1 GCES01105620 JAQ80702.1 PYGN01000313 PSN48697.1 GCES01046882 JAR39441.1 GCES01006847 JAR79476.1 GCES01019804 JAR66519.1 GCES01046574 JAR39749.1 GCES01108245 JAQ78077.1 GCES01119363 JAQ66959.1 KB201224 ESO98647.1 GCES01128106 JAQ58216.1 GCES01134318 JAQ52004.1 NHOQ01000736 PWA28943.1 GCES01129859 JAQ56463.1 CM000366 EDX17394.1 CH480859 EDW52748.1 BABH01037652 HADY01008008 HAEJ01012914 SBP46493.1 GCES01107100 JAQ79222.1 GCES01138729 JAQ47593.1
AGBW02010685 OWR48043.1 BABH01016863 JTDY01000233 KOB78143.1 ODYU01005946 SOQ47366.1 GAIX01003572 JAA88988.1 KQ459601 KPI93702.1 KQ459875 KPJ19633.1 PCG70576.1 KPJ19615.1 KPI93716.1 GDQN01008348 JAT82706.1 PCG70575.1 JTDY01008160 KOB64707.1 AGBW02011486 OWR46570.1 GDQN01002209 JAT88845.1 BABH01016826 ODYU01008528 SOQ52212.1 GDQN01003874 JAT87180.1 GAIX01003806 JAA88754.1 GDQN01010134 JAT80920.1 PCG70577.1 BABH01016827 JTDY01015286 KOB51959.1 GCES01066756 JAR19567.1 KOB64706.1 GCES01016586 JAR69737.1 GCES01145374 JAQ40948.1 GCES01140993 JAQ45329.1 CR790382 RJVU01040460 ROL46202.1 AGBW02008703 OWR52736.1 CH954180 EDV46831.1 AE014298 AGB95169.1 AY061144 AAF46335.1 AAL28692.1 CM000162 EDX02288.1 GECU01026801 JAS80905.1 HAEA01011435 SBQ39915.1 GBYX01226456 JAO70000.1 GCES01009514 JAR76809.1 HAEA01006085 SBQ34565.1 AYCK01020751 AYCK01020752 AYCK01020753 AYCK01020754 AYCK01019130 GAIX01009769 JAA82791.1 GCES01105620 JAQ80702.1 PYGN01000313 PSN48697.1 GCES01046882 JAR39441.1 GCES01006847 JAR79476.1 GCES01019804 JAR66519.1 GCES01046574 JAR39749.1 GCES01108245 JAQ78077.1 GCES01119363 JAQ66959.1 KB201224 ESO98647.1 GCES01128106 JAQ58216.1 GCES01134318 JAQ52004.1 NHOQ01000736 PWA28943.1 GCES01129859 JAQ56463.1 CM000366 EDX17394.1 CH480859 EDW52748.1 BABH01037652 HADY01008008 HAEJ01012914 SBP46493.1 GCES01107100 JAQ79222.1 GCES01138729 JAQ47593.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000037510
UP000053268
UP000053240
+ More
UP000261340 UP000265000 UP000000437 UP000261620 UP000261480 UP000008711 UP000000803 UP000192220 UP000264800 UP000002282 UP000261380 UP000261360 UP000261500 UP000002852 UP000028760 UP000261600 UP000264820 UP000261660 UP000245037 UP000261580 UP000030746 UP000192221 UP000000304 UP000001292 UP000265160 UP000261460 UP000264840
UP000261340 UP000265000 UP000000437 UP000261620 UP000261480 UP000008711 UP000000803 UP000192220 UP000264800 UP000002282 UP000261380 UP000261360 UP000261500 UP000002852 UP000028760 UP000261600 UP000264820 UP000261660 UP000245037 UP000261580 UP000030746 UP000192221 UP000000304 UP000001292 UP000265160 UP000261460 UP000264840
Interpro
ProteinModelPortal
A0A2H1VT86
A0A2A4JG74
A0A1E1WIZ6
A0A212F2U4
H9JMG1
A0A0L7LRQ9
+ More
A0A2H1W341 S4PGP2 A0A194PK55 A0A194RP61 A0A2A4JFN6 A0A194RQM6 A0A194PJV7 A0A1E1W6T3 A0A2A4JER5 A0A0L7KN53 A0A212EYL8 A0A1E1WPA8 H9JMG6 A0A2H1WH54 A0A1E1WJI4 S4PD46 A0A1E1W1P5 A0A2A4JEU2 H9JMG5 A0A0L7K2S7 A0A3Q0SQ85 A0A3Q2R090 A0A146VRK5 A0A0L7KNS3 A0A146ZUX2 A0A146PAU9 A0A146PMY2 A0A2R8Q0I5 A0A3Q3VV05 A0A3N0YIY5 A0A3B3XSK4 A0A212FG77 B3NX02 M9PHD3 Q9W3J2 A0A2I4BKY4 A0A3Q3AJH2 B4Q0F1 A0A2I4AJI5 A0A1B6I224 A0A3B5KUY2 A0A3Q3FVC2 A0A3B4Y2T3 A0A1A8DZZ5 A0A0S7JXW1 A0A147AE88 A0A2R8QH71 A0A3B3TK93 A0A3B5QTA6 A0A2I4BIT0 A0A1A8DJN5 A0A096M862 A0A096M2C9 A0A3Q3R3T5 S4NVL5 A0A3Q2PXS2 A0A146SJ19 A0A3Q3DHP3 A0A3Q3F3C3 A0A2P8YWR9 A0A087X5E0 A0A146XBG6 A0A3Q2UIJ1 A0A147ALR8 A0A3B3YA57 A0A3Q3A8R5 A0A146ZJI3 A0A146XDR0 A0A146SBU3 A0A146RF51 A0A087XBF4 A0A3Q4GBK2 A0A0G2KSV7 V4ANB0 A0A146QR30 A0A146Q7L4 A0A315W029 A0A1W4VWH2 A0A3Q2PS42 A0A3B5PVQ2 A0A146QK88 B4R6G4 A0A3Q2XZ96 A0A3B3UY54 B4IKX7 A0A3P9BXK5 H9J326 A0A3B3UEH5 A0A1A7ZVQ2 A0A146SEV3 A0A3B4H1W0 A0A146PUD5 A0A3Q3CRW7
A0A2H1W341 S4PGP2 A0A194PK55 A0A194RP61 A0A2A4JFN6 A0A194RQM6 A0A194PJV7 A0A1E1W6T3 A0A2A4JER5 A0A0L7KN53 A0A212EYL8 A0A1E1WPA8 H9JMG6 A0A2H1WH54 A0A1E1WJI4 S4PD46 A0A1E1W1P5 A0A2A4JEU2 H9JMG5 A0A0L7K2S7 A0A3Q0SQ85 A0A3Q2R090 A0A146VRK5 A0A0L7KNS3 A0A146ZUX2 A0A146PAU9 A0A146PMY2 A0A2R8Q0I5 A0A3Q3VV05 A0A3N0YIY5 A0A3B3XSK4 A0A212FG77 B3NX02 M9PHD3 Q9W3J2 A0A2I4BKY4 A0A3Q3AJH2 B4Q0F1 A0A2I4AJI5 A0A1B6I224 A0A3B5KUY2 A0A3Q3FVC2 A0A3B4Y2T3 A0A1A8DZZ5 A0A0S7JXW1 A0A147AE88 A0A2R8QH71 A0A3B3TK93 A0A3B5QTA6 A0A2I4BIT0 A0A1A8DJN5 A0A096M862 A0A096M2C9 A0A3Q3R3T5 S4NVL5 A0A3Q2PXS2 A0A146SJ19 A0A3Q3DHP3 A0A3Q3F3C3 A0A2P8YWR9 A0A087X5E0 A0A146XBG6 A0A3Q2UIJ1 A0A147ALR8 A0A3B3YA57 A0A3Q3A8R5 A0A146ZJI3 A0A146XDR0 A0A146SBU3 A0A146RF51 A0A087XBF4 A0A3Q4GBK2 A0A0G2KSV7 V4ANB0 A0A146QR30 A0A146Q7L4 A0A315W029 A0A1W4VWH2 A0A3Q2PS42 A0A3B5PVQ2 A0A146QK88 B4R6G4 A0A3Q2XZ96 A0A3B3UY54 B4IKX7 A0A3P9BXK5 H9J326 A0A3B3UEH5 A0A1A7ZVQ2 A0A146SEV3 A0A3B4H1W0 A0A146PUD5 A0A3Q3CRW7
PDB
5V3G
E-value=1.13503e-19,
Score=234
Ontologies
GO
Topology
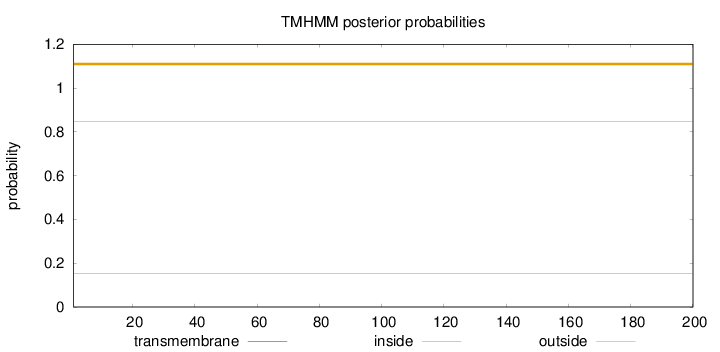
Length:
200
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15404
outside
1 - 200
Population Genetic Test Statistics
Pi
203.759059
Theta
188.195586
Tajima's D
0.260243
CLR
0
CSRT
0.446827658617069
Interpretation
Uncertain
