Gene
KWMTBOMO15604
Pre Gene Modal
BGIBMGA010836
Annotation
PREDICTED:_phosphatidylinositide_phosphatase_SAC2_isoform_X2_[Bombyx_mori]
Full name
Phosphatidylinositide phosphatase SAC2
Alternative Name
Inositol polyphosphate 5-phosphatase F
Sac domain-containing inositol phosphatase 2
Sac domain-containing phosphoinositide 4-phosphatase 2
Sac domain-containing inositol phosphatase 2
Sac domain-containing phosphoinositide 4-phosphatase 2
Location in the cell
Nuclear Reliability : 3.457
Sequence
CDS
ATGGAATTATTTCGTTCGGAACTCTATTACATTTTCGTCAGAAATGAGTCAAGTTTGTGGTGGAACAGATTATCGGGGGCTTTTTCTGTGCGATCAGCCTGGGACATGTCTGACATAGAAGATATAGAATGCCTCGGTATAACTGAAGGTATTATTGGAAAGGTCGAGCATTCCAATATATTCGAACCCCGCTTAATGATAATCAAAGAAAGTGCTCCCGTCGGCAACATATACTTTCATCACACCATTCATAAAGTAAAATCAATATGTTTTTTAAACATGGGCATAGCAAACCAAGAACTTGAACTGTGTCCTTGCACTAAACATGGCTCTTCCACATCTGTGCTGAGTTCTAATAAAAAAATAGGCAGTCGTCTGTTTGAAAATTCAGCATTTTTGAATAAAACTGTTGGAGCTGTAAAGAATGTAAGCAACACTATTAAAACAACTACACAGCAAGCAGCTACACAGGTGAAACAAACAGTAAAAAAGCAAAGAGATCCTAAAGTGGCAGAACGTTTTGAGAAGCGGCTCACGGATGAATTGCATAAAATCTTCGATGATTCACACAACTTTTATTATTCGAGAACATTGGACTTGACAAACAGCCTCCAAAGACAATATGAGATAGAAAAGATGTTGGAAACTGAAGAAGGTACCGGACGACCGGTCACAGACATAACGAGATGGTGGAAACATGTTGATGATAGATTCTTCTGGAATAAACATATGTTGAAAGATATTATTGCACTTGGGAGTCCAGATTGTGATCAGTGGATATTGCCAATAATTCAAGGATTCGTACATCTGTCTCAAATAGCCGTGGATCCAGCTGATTCTAACCCATTGAACACGGAATCTCTCACAAACACGAGCACATGTGATGAAACATTTACTCTCGGTTTAATATCCAGACGGTCAAGATATCAAGCTGGGACTCGTTACAATCGACGCGGTATCGAACAAAGCGGAAAAGTGGCTAATTATGTTGAAACCGAGCAAATTGTGTCGATAATTTGTTCGGATAGCATCCACAGGGCTTCATTTGTACAGGTACGAGGTTCGGTTCCAATCTTCTGGAGCCAGCCTGAATACAAGTTTCGTCCGCCACCACGACTTGATAAAACCGAAGAGGAGTCCCATGCCGCCTTCAAGAAGCATTTCGAAGAACAACTGAACACTTACAAGCAAGTGTGCATAGTGAACTTAGTCGAGCAGCAGGGCAGAGAACGCGTGATATGGGAGGCGTACGGCAACCACGTCCTCAAATACGACAACCCTAATCTCATATATGCGACCTTTGACTTCCACGAGTACTGCCGCGGTATGCACTACGAAAACGTGAGCATCCTCATCAACGCAATAGCGGACATAATAACCGACATGCGTTTCTGTTGGCGTGACGACCGCGGCGTTATATGCACGCAGAACGGCGTCTTCAGAGTCAACTGCATCGATTGTCTGGATAGAACGAACGTCGTACAAACTGCTATCGCTAAATACGTGTTGGAGTTGCAACTGTGCCGGCTGGGGTTGGGCACGCCTGGCTGTGGTCTGCCGCCGCCGCTTCAGAGGGCGTTCCTCACCATGTGGGCGGACAACGGCGATGTCGTGTCGAGGCAATACGCCGGAACGAAGGCCCTCAAGGGGGACTACACACGTACAGGCGAACGAAAGATCACGGGAATGATGAAGGACGGCGTTGCATCAGCAAATCGATATTATTTGTCAACTTTCAAGGATGCGCTGCGTCAAGTGGCCATAGACGTAATGACGGGCGAATCGAAAACGATACCGGAACAACTGATCGTGCCGGACTGCAGCCGATGCACTTCCGTTAAGGATCAAGCTGCCCCGGACACCGCTGCGATGGCGCAACACGTCAAGTCCCTAATCGACGACTGCAAGAAAATGTTGGTCGATACAGAACCGATACTTGGATCGTGGGGTCTGATCGACGCGGATCCACACACGGGCGATCCCCACGAAACGGAAATGGATAGCGTATTAGTTCTGACTAGTGAAGCCTACTACGTGGCGGACTACGACGAGACGTCGGACAGGCTGCTGTCCGTGCAGAGGGTGCCGCTGCGTGACGTCATCTCCATAGAGCTGGGAACGTTCGACGCAAGTTCCACTATTTTCGGCGTCGGTCGCAAGTCAAATTCGGAGACGGTATACTGCATGCGCATCAATTACAGTTACAACGGCGATAGTGGATACTTCCACATGTTCAGGTCGACCTCGCTTCGGTTCTTCAACAACATGGCGGTCGCAATCAACACCAGAGACGAGATGATAGAATCGCTGCACTCCATATGCGAATCCATGGTGGTGGCCCGTGAGGTCGCCAAGCTGCCGCCGATACCCTTCCACGATGGCATCAAATTGGAAAAGAGGAAATCGAAGGTACATCCGAATCACACTCAAGCCGGGGGAGCCCGTGCGTCTCCTTACCTGGACCTCTCTCGTCTGCCGACCCTCACCAGGAACGTCAGCGAGACGCAACTGGTGGCCGACATTCGGAGCGTCGGATCGAAGGCGTTCAACAACATGACGGAACAGTTCAGCAAACTAAACAAACTCAGTCACTCTCTGAACGCCAGAGCTAGACCGAGCCTCCAGCTAAAGTTCGACCAGAACACGACGAAAACGAAAAAATTATTCACGTTAGGTCAAAACAAAAGCGACAACAAGAAGAAGGGCAGCCTATCGGACGGCATGAGCTCCGATTACTCGTCGGACGATGAAAACCGGACCAACATATTCGAGCCGACCCTGGACAACTTCGAGAACACGCAGCACTACATCGGGAAACCGAACAGTTCGGGAAACGAAAACGACTGCGAACTTATCGAGAATCCACTGTATTCATCGAAAATCGAACCAAACGACAACCAAGAAGTCTTCATCGCGAATGAAACACCGCCCGCCGTCAACTTGCCGAAAACGCCGACTAATATCGGCAAAGTAAACCCGTTCAATTCGGAGAGGGCGCCCGAAATACAAATCGAGACGGAAGGTTCCGGAAAACCGGCACCACCGAACTCTCTGATGCTCTCGCAGAAGCTGTCCCACAGCTCGAACGATGTCAATTATGGGGAGAGCGATGGGGTCAATTACCACGTGAGGTCGAACTCCCAGCACGAGATCGTGCTGAATATCGCATCGTCCCACAGCGAGTCCGCTTTGAGGCACCTCAACTATATCTCCAGTCCAGTTTCCAGCACCAGCACGACCAAAGACCTGGTGCTTTCGCCCTTAACGAAGCTGGCTAAAGGAGTACAAAATCTTGGTGCTAATTTAGATCCGAGGAAGATAAAGGGTTCAGCGGCAGTTAAGCATGTAACGGAACAACAGTATGTCGAACATAGGAAGTTGCAGGAGAAATGGCGCGACTGTAACACCAGACTCGTTGCGTTGTAA
Protein
MELFRSELYYIFVRNESSLWWNRLSGAFSVRSAWDMSDIEDIECLGITEGIIGKVEHSNIFEPRLMIIKESAPVGNIYFHHTIHKVKSICFLNMGIANQELELCPCTKHGSSTSVLSSNKKIGSRLFENSAFLNKTVGAVKNVSNTIKTTTQQAATQVKQTVKKQRDPKVAERFEKRLTDELHKIFDDSHNFYYSRTLDLTNSLQRQYEIEKMLETEEGTGRPVTDITRWWKHVDDRFFWNKHMLKDIIALGSPDCDQWILPIIQGFVHLSQIAVDPADSNPLNTESLTNTSTCDETFTLGLISRRSRYQAGTRYNRRGIEQSGKVANYVETEQIVSIICSDSIHRASFVQVRGSVPIFWSQPEYKFRPPPRLDKTEEESHAAFKKHFEEQLNTYKQVCIVNLVEQQGRERVIWEAYGNHVLKYDNPNLIYATFDFHEYCRGMHYENVSILINAIADIITDMRFCWRDDRGVICTQNGVFRVNCIDCLDRTNVVQTAIAKYVLELQLCRLGLGTPGCGLPPPLQRAFLTMWADNGDVVSRQYAGTKALKGDYTRTGERKITGMMKDGVASANRYYLSTFKDALRQVAIDVMTGESKTIPEQLIVPDCSRCTSVKDQAAPDTAAMAQHVKSLIDDCKKMLVDTEPILGSWGLIDADPHTGDPHETEMDSVLVLTSEAYYVADYDETSDRLLSVQRVPLRDVISIELGTFDASSTIFGVGRKSNSETVYCMRINYSYNGDSGYFHMFRSTSLRFFNNMAVAINTRDEMIESLHSICESMVVAREVAKLPPIPFHDGIKLEKRKSKVHPNHTQAGGARASPYLDLSRLPTLTRNVSETQLVADIRSVGSKAFNNMTEQFSKLNKLSHSLNARARPSLQLKFDQNTTKTKKLFTLGQNKSDNKKKGSLSDGMSSDYSSDDENRTNIFEPTLDNFENTQHYIGKPNSSGNENDCELIENPLYSSKIEPNDNQEVFIANETPPAVNLPKTPTNIGKVNPFNSERAPEIQIETEGSGKPAPPNSLMLSQKLSHSSNDVNYGESDGVNYHVRSNSQHEIVLNIASSHSESALRHLNYISSPVSSTSTTKDLVLSPLTKLAKGVQNLGANLDPRKIKGSAAVKHVTEQQYVEHRKLQEKWRDCNTRLVAL
Summary
Description
Inositol 4-phosphatase which mainly acts on phosphatidylinositol 4-phosphate. May be functionally linked to OCRL, which converts phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol, for a sequential dephosphorylation of phosphatidylinositol 4,5-bisphosphate at the 5 and 4 position of inositol, thus playing an important role in the endocytic recycling.
Catalytic Activity
a myo-inositol phosphate + H2O = myo-inositol + phosphate
Keywords
Coated pit
Complete proteome
Endosome
Hydrolase
Membrane
Reference proteome
Feature
chain Phosphatidylinositide phosphatase SAC2
Uniprot
A0A2A4JFU2
A0A212EYN3
A0A194RQD6
A0A1Y1MJE7
A0A310SP49
A0A0C9RM71
+ More
A0A067R946 K7IP27 E0VJW7 W8BDH9 A0A336M9Y8 A0A1I8M1E0 Q16HF8 A0A182J428 A0A1I8Q0N9 A0A034VS64 A0A1B6C6E6 A0A0Q9WTV4 A0A131Y988 A0A146LP15 V5IIQ7 A0A0Q9XAI7 A0A0K8U290 A0A084VQC4 A0A1Q3FAY8 B0WSI4 E1JIS1 W8BIC1 A0A1W4VZ32 Q7QFH3 A0A023EYP6 A0A0A1X3H9 A0A0A1XQM6 A0A0Q9X481 W5JX57 A0A0R1E1L1 A0A224X6C7 Q59DV6 A0A3B0K3W3 Q9VD98 Q8MRH8 A0A0A1X9W4 V5HYT9 A0A0A1XL36 B4R0E8 I5ANH5 A0A224Z5A0 A0A034VUT2 A0A0K8UM12 A0A131YT97 B3LVD2 A0A2M4A9F2 A0A0P4WT57 A0A0T6B660 E9HJ60 A0A0P5ZVW9 A0A0N8B491 T1KD31 A0A0P6G8E8 A0A0P5LD57 A0A0P5S5B5 A0A0P5HK48 A0A0P5KJA7 A0A0P6IFE7 A0A218UQG6 A0A3N0YTP4 F1NY27 H0ZLQ0 A0A3B1IRX4 A8E7C5 A0A250Y6W6 A0A2I4D5E7 A0A2K5PBY2 A0A3Q3MFX2 A0A2K6QFH9 A0A2K5CI37
A0A067R946 K7IP27 E0VJW7 W8BDH9 A0A336M9Y8 A0A1I8M1E0 Q16HF8 A0A182J428 A0A1I8Q0N9 A0A034VS64 A0A1B6C6E6 A0A0Q9WTV4 A0A131Y988 A0A146LP15 V5IIQ7 A0A0Q9XAI7 A0A0K8U290 A0A084VQC4 A0A1Q3FAY8 B0WSI4 E1JIS1 W8BIC1 A0A1W4VZ32 Q7QFH3 A0A023EYP6 A0A0A1X3H9 A0A0A1XQM6 A0A0Q9X481 W5JX57 A0A0R1E1L1 A0A224X6C7 Q59DV6 A0A3B0K3W3 Q9VD98 Q8MRH8 A0A0A1X9W4 V5HYT9 A0A0A1XL36 B4R0E8 I5ANH5 A0A224Z5A0 A0A034VUT2 A0A0K8UM12 A0A131YT97 B3LVD2 A0A2M4A9F2 A0A0P4WT57 A0A0T6B660 E9HJ60 A0A0P5ZVW9 A0A0N8B491 T1KD31 A0A0P6G8E8 A0A0P5LD57 A0A0P5S5B5 A0A0P5HK48 A0A0P5KJA7 A0A0P6IFE7 A0A218UQG6 A0A3N0YTP4 F1NY27 H0ZLQ0 A0A3B1IRX4 A8E7C5 A0A250Y6W6 A0A2I4D5E7 A0A2K5PBY2 A0A3Q3MFX2 A0A2K6QFH9 A0A2K5CI37
EC Number
3.1.3.25
Pubmed
22118469
26354079
28004739
24845553
20075255
20566863
+ More
24495485 25315136 17510324 25348373 17994087 26823975 25765539 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12364791 25474469 25830018 20920257 23761445 17550304 26109357 26109356 15632085 28797301 26830274 21292972 15592404 20360741 25329095 23594743 28087693 25362486
24495485 25315136 17510324 25348373 17994087 26823975 25765539 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12364791 25474469 25830018 20920257 23761445 17550304 26109357 26109356 15632085 28797301 26830274 21292972 15592404 20360741 25329095 23594743 28087693 25362486
EMBL
NWSH01001646
PCG70578.1
AGBW02011486
OWR46571.1
KQ459875
KPJ19614.1
+ More
GEZM01030106 JAV85803.1 KQ760549 OAD59920.1 GBYB01014482 JAG84249.1 KK852660 KDR19126.1 DS235231 EEB13673.1 GAMC01009808 JAB96747.1 UFQT01000610 SSX25709.1 CH478175 EAT33690.1 GAKP01013663 JAC45289.1 GEDC01028429 JAS08869.1 CH940650 KRF83573.1 GEFM01000359 JAP75437.1 GDHC01010499 JAQ08130.1 GANP01002767 JAB81701.1 CH933806 KRG01537.1 GDHF01031455 JAI20859.1 ATLV01015187 KE525003 KFB40168.1 GFDL01010309 JAV24736.1 DS232072 EDS33914.1 AE014297 ACZ94971.1 GAMC01009807 JAB96748.1 AAAB01008846 EAA06429.4 GBBI01004450 JAC14262.1 GBXI01008418 JAD05874.1 GBXI01001030 JAD13262.1 CH964251 KRF99757.1 ADMH02000018 ETN68075.1 CM000160 KRK03049.1 GFTR01008399 JAW08027.1 AAX52970.3 OUUW01000005 SPP80679.1 BT044234 AAF55899.2 ACH92299.1 AY119613 AAM50267.1 GBXI01006772 JAD07520.1 GANP01002766 JAB81702.1 GBXI01002218 JAD12074.1 CM000364 EDX12073.1 CM000070 EIM52510.1 GFPF01012163 MAA23309.1 GAKP01013664 JAC45288.1 GDHF01024766 JAI27548.1 GEDV01006785 JAP81772.1 CH902617 EDV43664.2 GGFK01004096 MBW37417.1 GDRN01063020 JAI65019.1 LJIG01009806 KRT82343.1 GL732660 EFX68224.1 GDIP01051236 LRGB01002190 JAM52479.1 KZS08589.1 GDIQ01214331 JAK37394.1 CAEY01002009 GDIQ01039794 JAN54943.1 GDIQ01171447 JAK80278.1 GDIQ01101082 JAL50644.1 GDIQ01226982 JAK24743.1 GDIQ01183292 JAK68433.1 GDIQ01005319 JAN89418.1 MUZQ01000180 OWK55979.1 RJVU01026577 ROL49564.1 AADN05000306 ABQF01046771 BX005252 BX530079 GFFV01000584 JAV39361.1
GEZM01030106 JAV85803.1 KQ760549 OAD59920.1 GBYB01014482 JAG84249.1 KK852660 KDR19126.1 DS235231 EEB13673.1 GAMC01009808 JAB96747.1 UFQT01000610 SSX25709.1 CH478175 EAT33690.1 GAKP01013663 JAC45289.1 GEDC01028429 JAS08869.1 CH940650 KRF83573.1 GEFM01000359 JAP75437.1 GDHC01010499 JAQ08130.1 GANP01002767 JAB81701.1 CH933806 KRG01537.1 GDHF01031455 JAI20859.1 ATLV01015187 KE525003 KFB40168.1 GFDL01010309 JAV24736.1 DS232072 EDS33914.1 AE014297 ACZ94971.1 GAMC01009807 JAB96748.1 AAAB01008846 EAA06429.4 GBBI01004450 JAC14262.1 GBXI01008418 JAD05874.1 GBXI01001030 JAD13262.1 CH964251 KRF99757.1 ADMH02000018 ETN68075.1 CM000160 KRK03049.1 GFTR01008399 JAW08027.1 AAX52970.3 OUUW01000005 SPP80679.1 BT044234 AAF55899.2 ACH92299.1 AY119613 AAM50267.1 GBXI01006772 JAD07520.1 GANP01002766 JAB81702.1 GBXI01002218 JAD12074.1 CM000364 EDX12073.1 CM000070 EIM52510.1 GFPF01012163 MAA23309.1 GAKP01013664 JAC45288.1 GDHF01024766 JAI27548.1 GEDV01006785 JAP81772.1 CH902617 EDV43664.2 GGFK01004096 MBW37417.1 GDRN01063020 JAI65019.1 LJIG01009806 KRT82343.1 GL732660 EFX68224.1 GDIP01051236 LRGB01002190 JAM52479.1 KZS08589.1 GDIQ01214331 JAK37394.1 CAEY01002009 GDIQ01039794 JAN54943.1 GDIQ01171447 JAK80278.1 GDIQ01101082 JAL50644.1 GDIQ01226982 JAK24743.1 GDIQ01183292 JAK68433.1 GDIQ01005319 JAN89418.1 MUZQ01000180 OWK55979.1 RJVU01026577 ROL49564.1 AADN05000306 ABQF01046771 BX005252 BX530079 GFFV01000584 JAV39361.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000027135
UP000002358
UP000009046
+ More
UP000095301 UP000008820 UP000075880 UP000095300 UP000008792 UP000009192 UP000030765 UP000002320 UP000000803 UP000192221 UP000007062 UP000007798 UP000000673 UP000002282 UP000268350 UP000000304 UP000001819 UP000007801 UP000000305 UP000076858 UP000015104 UP000197619 UP000000539 UP000007754 UP000018467 UP000000437 UP000192220 UP000233040 UP000261640 UP000233200 UP000233020
UP000095301 UP000008820 UP000075880 UP000095300 UP000008792 UP000009192 UP000030765 UP000002320 UP000000803 UP000192221 UP000007062 UP000007798 UP000000673 UP000002282 UP000268350 UP000000304 UP000001819 UP000007801 UP000000305 UP000076858 UP000015104 UP000197619 UP000000539 UP000007754 UP000018467 UP000000437 UP000192220 UP000233040 UP000261640 UP000233200 UP000233020
ProteinModelPortal
A0A2A4JFU2
A0A212EYN3
A0A194RQD6
A0A1Y1MJE7
A0A310SP49
A0A0C9RM71
+ More
A0A067R946 K7IP27 E0VJW7 W8BDH9 A0A336M9Y8 A0A1I8M1E0 Q16HF8 A0A182J428 A0A1I8Q0N9 A0A034VS64 A0A1B6C6E6 A0A0Q9WTV4 A0A131Y988 A0A146LP15 V5IIQ7 A0A0Q9XAI7 A0A0K8U290 A0A084VQC4 A0A1Q3FAY8 B0WSI4 E1JIS1 W8BIC1 A0A1W4VZ32 Q7QFH3 A0A023EYP6 A0A0A1X3H9 A0A0A1XQM6 A0A0Q9X481 W5JX57 A0A0R1E1L1 A0A224X6C7 Q59DV6 A0A3B0K3W3 Q9VD98 Q8MRH8 A0A0A1X9W4 V5HYT9 A0A0A1XL36 B4R0E8 I5ANH5 A0A224Z5A0 A0A034VUT2 A0A0K8UM12 A0A131YT97 B3LVD2 A0A2M4A9F2 A0A0P4WT57 A0A0T6B660 E9HJ60 A0A0P5ZVW9 A0A0N8B491 T1KD31 A0A0P6G8E8 A0A0P5LD57 A0A0P5S5B5 A0A0P5HK48 A0A0P5KJA7 A0A0P6IFE7 A0A218UQG6 A0A3N0YTP4 F1NY27 H0ZLQ0 A0A3B1IRX4 A8E7C5 A0A250Y6W6 A0A2I4D5E7 A0A2K5PBY2 A0A3Q3MFX2 A0A2K6QFH9 A0A2K5CI37
A0A067R946 K7IP27 E0VJW7 W8BDH9 A0A336M9Y8 A0A1I8M1E0 Q16HF8 A0A182J428 A0A1I8Q0N9 A0A034VS64 A0A1B6C6E6 A0A0Q9WTV4 A0A131Y988 A0A146LP15 V5IIQ7 A0A0Q9XAI7 A0A0K8U290 A0A084VQC4 A0A1Q3FAY8 B0WSI4 E1JIS1 W8BIC1 A0A1W4VZ32 Q7QFH3 A0A023EYP6 A0A0A1X3H9 A0A0A1XQM6 A0A0Q9X481 W5JX57 A0A0R1E1L1 A0A224X6C7 Q59DV6 A0A3B0K3W3 Q9VD98 Q8MRH8 A0A0A1X9W4 V5HYT9 A0A0A1XL36 B4R0E8 I5ANH5 A0A224Z5A0 A0A034VUT2 A0A0K8UM12 A0A131YT97 B3LVD2 A0A2M4A9F2 A0A0P4WT57 A0A0T6B660 E9HJ60 A0A0P5ZVW9 A0A0N8B491 T1KD31 A0A0P6G8E8 A0A0P5LD57 A0A0P5S5B5 A0A0P5HK48 A0A0P5KJA7 A0A0P6IFE7 A0A218UQG6 A0A3N0YTP4 F1NY27 H0ZLQ0 A0A3B1IRX4 A8E7C5 A0A250Y6W6 A0A2I4D5E7 A0A2K5PBY2 A0A3Q3MFX2 A0A2K6QFH9 A0A2K5CI37
PDB
4TU3
E-value=3.41362e-64,
Score=626
Ontologies
PATHWAY
GO
GO:0042578
GO:0045334
GO:0046856
GO:0005769
GO:2001135
GO:0043812
GO:0001921
GO:0005905
GO:0048015
GO:0043025
GO:0052833
GO:2000145
GO:0034596
GO:0008344
GO:0042532
GO:0055037
GO:0014898
GO:0034595
GO:0030424
GO:0030425
GO:0048681
GO:0072583
GO:0042803
GO:0031161
GO:0008934
GO:0052832
GO:0003676
GO:0005634
GO:0008270
GO:0030126
GO:0030117
GO:0006418
GO:0003707
GO:0006281
GO:0006259
Topology
Subcellular location
Membrane
Also found on macropinosomes. With evidence from 1 publications.
Clathrin-coated pit Also found on macropinosomes. With evidence from 1 publications.
Early endosome Also found on macropinosomes. With evidence from 1 publications.
Recycling endosome Also found on macropinosomes. With evidence from 1 publications.
Clathrin-coated pit Also found on macropinosomes. With evidence from 1 publications.
Early endosome Also found on macropinosomes. With evidence from 1 publications.
Recycling endosome Also found on macropinosomes. With evidence from 1 publications.
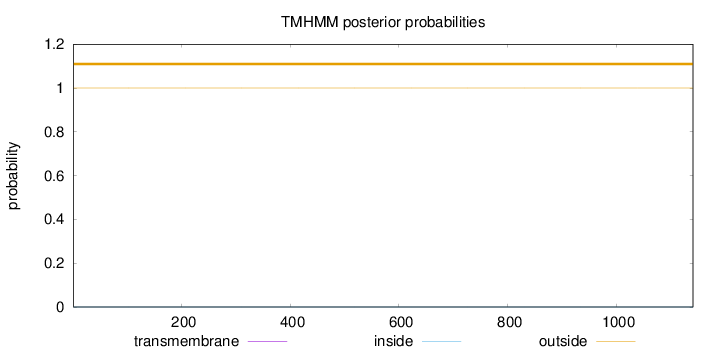
Length:
1141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000460000000000001
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00002
outside
1 - 1141
Population Genetic Test Statistics
Pi
28.720354
Theta
27.35354
Tajima's D
-1.874485
CLR
1.596886
CSRT
0.0223988800559972
Interpretation
Possibly Positive selection
