Gene
KWMTBOMO15596
Pre Gene Modal
BGIBMGA010722
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.866
Sequence
CDS
ATGTCAATGGGATGGACATCACCAGTGTTGCCACAGTTGCGAGGACCCAACTCCCCACTGCCAGAACAGCCTACGCTACAGCAAGAATCCTGGATCGGCTCTCTGCTCGTTCTCGGCGGATTGATCGGCCCATTCATCACAGTGCCATTGTCAAAGCACGTTGGCCGGAGATGGCTGATTATGGGATCGAACGTGCCCCTACTACTGGGCTGGTTGCTCGCCGGAGTCACAACTAACTTGCCAACGTTGTACGTCGCGAGGTTGGCCTGGGGCTGTGCTACTGCCATGCAATTTGCGACAGTACCTATTTACATAGGAGAAATTGCCGAGGACAAAATCCGGGGAGCTCTGAGCGCGTTTTTCTTGTTGTTCATCAATATCGGCTTTTTGTTGGCCTACGCCATCGGTCCTTTCACCAGCTATTGGGGCCTCACAGCCTGTGGTGGAATTATATCCCTGTTATATCTACCGTTTTCTTGGTATTTACCAGAAACACCGTTCTTTTTGGTGTTCAAAGGTTAG
Protein
MSMGWTSPVLPQLRGPNSPLPEQPTLQQESWIGSLLVLGGLIGPFITVPLSKHVGRRWLIMGSNVPLLLGWLLAGVTTNLPTLYVARLAWGCATAMQFATVPIYIGEIAEDKIRGALSAFFLLFINIGFLLAYAIGPFTSYWGLTACGGIISLLYLPFSWYLPETPFFLVFKG
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JMG9
A0A0N0PA76
A0A2A4JTW5
A0A212EIU1
A0A3S2LU80
A0A2H1VN09
+ More
A0A194RQD1 H9JMJ0 W8B3Q4 A0A2A4JNK2 W8BDP5 A0A2A4JQ62 B4JH52 A0A2H1VKV6 A0A194PK39 B4LXB3 A0A195CMH5 A0A1L8EH92 A0A1L8EF30 A0A0M5JD36 H9JMI8 B4NJT9 B4KA40 A0A2W1BVC4 A0A3L8DXN6 E9IQR3 E2B123 A0A0K8W987 A0A2H1VF40 A0A0K8U506 A0A1I8P6M8 A0A0K8WL46 A8CWB1 A0A151WM18 A0A2A4JTT1 H9JMI9 A0A1I8MD33 A0A0N0BDQ1 A0A084WNA9 B0X5T8 A0A195EU19 T1PBE5 A0A1I8NJI3 A0A1L8DIL8 A0A1L8DIL5 F4WP33 A0A0C9R330 A0A1L8DIJ3 A0A1L8DIK8 A0A034W7W8 A0A182IRD4 U5ESS0 A0A1B0D4P3 A0A182S4P1 A0A212ERK6 B4PVJ3 A0A1I8MXU6 A0A0L0CG94 B4QXL4 A0A3B0K5K3 B4I487 Q7JVN6 A0A3B0JXL8 D6X0L8 A0A195B938 A0A158NGK6 A0A2M4A0H9 A0A2M4A0I2 K7IP31 A0A194PXP0 A0A182XYW6 A0A0J7NVW1 B4G348 A0A232F7N7 B0X5T9 A0A182JUH7 A0A182WJ81 E2BLV7 A0A182J5I6 A0A2A3E9H1 B3NYI2 W5JRP7 A0A336M0J9 Q297W3 W4VS73 A0A0A1WV08 A0A084VDW1 A0A1I8MD12 A0A182NHV2 A0A1W4UWZ5 A0A0A1WDI7 A0A182LZZ1 A0A182QUI1 A0A182QHE6 S4P5I1 A0A182RSN7 A0A1S4FT34 Q16Q39 W5J543
A0A194RQD1 H9JMJ0 W8B3Q4 A0A2A4JNK2 W8BDP5 A0A2A4JQ62 B4JH52 A0A2H1VKV6 A0A194PK39 B4LXB3 A0A195CMH5 A0A1L8EH92 A0A1L8EF30 A0A0M5JD36 H9JMI8 B4NJT9 B4KA40 A0A2W1BVC4 A0A3L8DXN6 E9IQR3 E2B123 A0A0K8W987 A0A2H1VF40 A0A0K8U506 A0A1I8P6M8 A0A0K8WL46 A8CWB1 A0A151WM18 A0A2A4JTT1 H9JMI9 A0A1I8MD33 A0A0N0BDQ1 A0A084WNA9 B0X5T8 A0A195EU19 T1PBE5 A0A1I8NJI3 A0A1L8DIL8 A0A1L8DIL5 F4WP33 A0A0C9R330 A0A1L8DIJ3 A0A1L8DIK8 A0A034W7W8 A0A182IRD4 U5ESS0 A0A1B0D4P3 A0A182S4P1 A0A212ERK6 B4PVJ3 A0A1I8MXU6 A0A0L0CG94 B4QXL4 A0A3B0K5K3 B4I487 Q7JVN6 A0A3B0JXL8 D6X0L8 A0A195B938 A0A158NGK6 A0A2M4A0H9 A0A2M4A0I2 K7IP31 A0A194PXP0 A0A182XYW6 A0A0J7NVW1 B4G348 A0A232F7N7 B0X5T9 A0A182JUH7 A0A182WJ81 E2BLV7 A0A182J5I6 A0A2A3E9H1 B3NYI2 W5JRP7 A0A336M0J9 Q297W3 W4VS73 A0A0A1WV08 A0A084VDW1 A0A1I8MD12 A0A182NHV2 A0A1W4UWZ5 A0A0A1WDI7 A0A182LZZ1 A0A182QUI1 A0A182QHE6 S4P5I1 A0A182RSN7 A0A1S4FT34 Q16Q39 W5J543
Pubmed
19121390
26354079
22118469
24495485
17994087
18057021
+ More
28756777 30249741 21282665 20798317 18194529 25315136 24438588 21719571 25348373 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 21347285 20075255 25244985 28648823 20920257 23761445 15632085 23185243 24391959 25830018 23622113 17510324
28756777 30249741 21282665 20798317 18194529 25315136 24438588 21719571 25348373 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 21347285 20075255 25244985 28648823 20920257 23761445 15632085 23185243 24391959 25830018 23622113 17510324
EMBL
BABH01016814
KQ459078
KPJ03826.1
NWSH01000595
PCG75451.1
AGBW02014588
+ More
OWR41380.1 RSAL01000221 RVE44063.1 ODYU01003431 SOQ42187.1 KQ459875 KPJ19609.1 BABH01016691 BABH01016692 BABH01016693 BABH01016694 GAMC01018654 JAB87901.1 NWSH01000916 PCG73551.1 GAMC01018656 JAB87899.1 PCG73552.1 CH916369 EDV92743.1 ODYU01003112 SOQ41455.1 KQ459601 KPI93687.1 CH940650 EDW67791.1 KRF83468.1 KRF83469.1 KQ977574 KYN01895.1 GFDG01000707 JAV18092.1 GFDG01001479 JAV17320.1 CP012526 ALC47794.1 BABH01016697 CH964272 EDW85051.1 KRG00175.1 CH933806 EDW16715.2 KRG02269.1 KRG02270.1 KZ149936 PZC77147.1 QOIP01000003 RLU25240.1 GL764900 EFZ17069.1 GL444771 EFN60596.1 GDHF01004672 JAI47642.1 ODYU01001965 SOQ38884.1 GDHF01030597 JAI21717.1 GDHF01000517 JAI51797.1 EU124608 ABV60326.1 KQ982944 KYQ48914.1 NWSH01000598 PCG75421.1 BABH01016696 KQ435851 KOX70761.1 ATLV01024580 KE525352 KFB51703.1 DS232396 EDS41069.1 KQ981965 KYN31745.1 KA646086 AFP60715.1 GFDF01007791 JAV06293.1 GFDF01007792 JAV06292.1 GL888243 EGI63964.1 GBYB01002345 JAG72112.1 GFDF01007785 JAV06299.1 GFDF01007786 JAV06298.1 GAKP01008752 GAKP01008751 JAC50200.1 GANO01003127 JAB56744.1 AJVK01011560 AGBW02012985 OWR44136.1 CM000160 EDW97802.1 KRK03978.1 KRK03979.1 JRES01000423 KNC31408.1 CM000364 EDX11800.1 OUUW01000005 SPP80251.1 CH480821 EDW55030.1 AE014297 AY118796 AAF51941.1 AAM50656.1 SPP80250.1 KQ971371 EFA10112.2 KQ976542 KYM81051.1 ADTU01015114 GGFK01000993 MBW34314.1 GGFK01000934 MBW34255.1 KQ459589 KPI97524.1 LBMM01001313 KMQ96530.1 CH479179 EDW24243.1 NNAY01000793 OXU26469.1 EDS41070.1 GL449042 EFN83323.1 KZ288338 PBC27839.1 CH954181 EDV47963.1 ADMH02000549 ETN65958.1 UFQT01000389 SSX23856.1 CM000070 EAL28092.1 KRT00434.1 GAPU01000032 JAB84596.1 GBXI01011595 GBXI01005185 JAD02697.1 JAD09107.1 ATLV01011809 KE524723 KFB36155.1 GBXI01017546 JAC96745.1 AXCM01019063 AXCN02000379 GAIX01010740 JAA81820.1 CH477763 EAT36506.1 ADMH02002130 ETN58528.1
OWR41380.1 RSAL01000221 RVE44063.1 ODYU01003431 SOQ42187.1 KQ459875 KPJ19609.1 BABH01016691 BABH01016692 BABH01016693 BABH01016694 GAMC01018654 JAB87901.1 NWSH01000916 PCG73551.1 GAMC01018656 JAB87899.1 PCG73552.1 CH916369 EDV92743.1 ODYU01003112 SOQ41455.1 KQ459601 KPI93687.1 CH940650 EDW67791.1 KRF83468.1 KRF83469.1 KQ977574 KYN01895.1 GFDG01000707 JAV18092.1 GFDG01001479 JAV17320.1 CP012526 ALC47794.1 BABH01016697 CH964272 EDW85051.1 KRG00175.1 CH933806 EDW16715.2 KRG02269.1 KRG02270.1 KZ149936 PZC77147.1 QOIP01000003 RLU25240.1 GL764900 EFZ17069.1 GL444771 EFN60596.1 GDHF01004672 JAI47642.1 ODYU01001965 SOQ38884.1 GDHF01030597 JAI21717.1 GDHF01000517 JAI51797.1 EU124608 ABV60326.1 KQ982944 KYQ48914.1 NWSH01000598 PCG75421.1 BABH01016696 KQ435851 KOX70761.1 ATLV01024580 KE525352 KFB51703.1 DS232396 EDS41069.1 KQ981965 KYN31745.1 KA646086 AFP60715.1 GFDF01007791 JAV06293.1 GFDF01007792 JAV06292.1 GL888243 EGI63964.1 GBYB01002345 JAG72112.1 GFDF01007785 JAV06299.1 GFDF01007786 JAV06298.1 GAKP01008752 GAKP01008751 JAC50200.1 GANO01003127 JAB56744.1 AJVK01011560 AGBW02012985 OWR44136.1 CM000160 EDW97802.1 KRK03978.1 KRK03979.1 JRES01000423 KNC31408.1 CM000364 EDX11800.1 OUUW01000005 SPP80251.1 CH480821 EDW55030.1 AE014297 AY118796 AAF51941.1 AAM50656.1 SPP80250.1 KQ971371 EFA10112.2 KQ976542 KYM81051.1 ADTU01015114 GGFK01000993 MBW34314.1 GGFK01000934 MBW34255.1 KQ459589 KPI97524.1 LBMM01001313 KMQ96530.1 CH479179 EDW24243.1 NNAY01000793 OXU26469.1 EDS41070.1 GL449042 EFN83323.1 KZ288338 PBC27839.1 CH954181 EDV47963.1 ADMH02000549 ETN65958.1 UFQT01000389 SSX23856.1 CM000070 EAL28092.1 KRT00434.1 GAPU01000032 JAB84596.1 GBXI01011595 GBXI01005185 JAD02697.1 JAD09107.1 ATLV01011809 KE524723 KFB36155.1 GBXI01017546 JAC96745.1 AXCM01019063 AXCN02000379 GAIX01010740 JAA81820.1 CH477763 EAT36506.1 ADMH02002130 ETN58528.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000283053
UP000053240
+ More
UP000001070 UP000008792 UP000078542 UP000092553 UP000007798 UP000009192 UP000279307 UP000000311 UP000095300 UP000075809 UP000095301 UP000053105 UP000030765 UP000002320 UP000078541 UP000007755 UP000075880 UP000092462 UP000075900 UP000002282 UP000037069 UP000000304 UP000268350 UP000001292 UP000000803 UP000007266 UP000078540 UP000005205 UP000002358 UP000076408 UP000036403 UP000008744 UP000215335 UP000075881 UP000075920 UP000008237 UP000242457 UP000008711 UP000000673 UP000001819 UP000075884 UP000192221 UP000075883 UP000075886 UP000008820
UP000001070 UP000008792 UP000078542 UP000092553 UP000007798 UP000009192 UP000279307 UP000000311 UP000095300 UP000075809 UP000095301 UP000053105 UP000030765 UP000002320 UP000078541 UP000007755 UP000075880 UP000092462 UP000075900 UP000002282 UP000037069 UP000000304 UP000268350 UP000001292 UP000000803 UP000007266 UP000078540 UP000005205 UP000002358 UP000076408 UP000036403 UP000008744 UP000215335 UP000075881 UP000075920 UP000008237 UP000242457 UP000008711 UP000000673 UP000001819 UP000075884 UP000192221 UP000075883 UP000075886 UP000008820
Interpro
CDD
ProteinModelPortal
H9JMG9
A0A0N0PA76
A0A2A4JTW5
A0A212EIU1
A0A3S2LU80
A0A2H1VN09
+ More
A0A194RQD1 H9JMJ0 W8B3Q4 A0A2A4JNK2 W8BDP5 A0A2A4JQ62 B4JH52 A0A2H1VKV6 A0A194PK39 B4LXB3 A0A195CMH5 A0A1L8EH92 A0A1L8EF30 A0A0M5JD36 H9JMI8 B4NJT9 B4KA40 A0A2W1BVC4 A0A3L8DXN6 E9IQR3 E2B123 A0A0K8W987 A0A2H1VF40 A0A0K8U506 A0A1I8P6M8 A0A0K8WL46 A8CWB1 A0A151WM18 A0A2A4JTT1 H9JMI9 A0A1I8MD33 A0A0N0BDQ1 A0A084WNA9 B0X5T8 A0A195EU19 T1PBE5 A0A1I8NJI3 A0A1L8DIL8 A0A1L8DIL5 F4WP33 A0A0C9R330 A0A1L8DIJ3 A0A1L8DIK8 A0A034W7W8 A0A182IRD4 U5ESS0 A0A1B0D4P3 A0A182S4P1 A0A212ERK6 B4PVJ3 A0A1I8MXU6 A0A0L0CG94 B4QXL4 A0A3B0K5K3 B4I487 Q7JVN6 A0A3B0JXL8 D6X0L8 A0A195B938 A0A158NGK6 A0A2M4A0H9 A0A2M4A0I2 K7IP31 A0A194PXP0 A0A182XYW6 A0A0J7NVW1 B4G348 A0A232F7N7 B0X5T9 A0A182JUH7 A0A182WJ81 E2BLV7 A0A182J5I6 A0A2A3E9H1 B3NYI2 W5JRP7 A0A336M0J9 Q297W3 W4VS73 A0A0A1WV08 A0A084VDW1 A0A1I8MD12 A0A182NHV2 A0A1W4UWZ5 A0A0A1WDI7 A0A182LZZ1 A0A182QUI1 A0A182QHE6 S4P5I1 A0A182RSN7 A0A1S4FT34 Q16Q39 W5J543
A0A194RQD1 H9JMJ0 W8B3Q4 A0A2A4JNK2 W8BDP5 A0A2A4JQ62 B4JH52 A0A2H1VKV6 A0A194PK39 B4LXB3 A0A195CMH5 A0A1L8EH92 A0A1L8EF30 A0A0M5JD36 H9JMI8 B4NJT9 B4KA40 A0A2W1BVC4 A0A3L8DXN6 E9IQR3 E2B123 A0A0K8W987 A0A2H1VF40 A0A0K8U506 A0A1I8P6M8 A0A0K8WL46 A8CWB1 A0A151WM18 A0A2A4JTT1 H9JMI9 A0A1I8MD33 A0A0N0BDQ1 A0A084WNA9 B0X5T8 A0A195EU19 T1PBE5 A0A1I8NJI3 A0A1L8DIL8 A0A1L8DIL5 F4WP33 A0A0C9R330 A0A1L8DIJ3 A0A1L8DIK8 A0A034W7W8 A0A182IRD4 U5ESS0 A0A1B0D4P3 A0A182S4P1 A0A212ERK6 B4PVJ3 A0A1I8MXU6 A0A0L0CG94 B4QXL4 A0A3B0K5K3 B4I487 Q7JVN6 A0A3B0JXL8 D6X0L8 A0A195B938 A0A158NGK6 A0A2M4A0H9 A0A2M4A0I2 K7IP31 A0A194PXP0 A0A182XYW6 A0A0J7NVW1 B4G348 A0A232F7N7 B0X5T9 A0A182JUH7 A0A182WJ81 E2BLV7 A0A182J5I6 A0A2A3E9H1 B3NYI2 W5JRP7 A0A336M0J9 Q297W3 W4VS73 A0A0A1WV08 A0A084VDW1 A0A1I8MD12 A0A182NHV2 A0A1W4UWZ5 A0A0A1WDI7 A0A182LZZ1 A0A182QUI1 A0A182QHE6 S4P5I1 A0A182RSN7 A0A1S4FT34 Q16Q39 W5J543
PDB
4LDS
E-value=6.08599e-08,
Score=131
Ontologies
GO
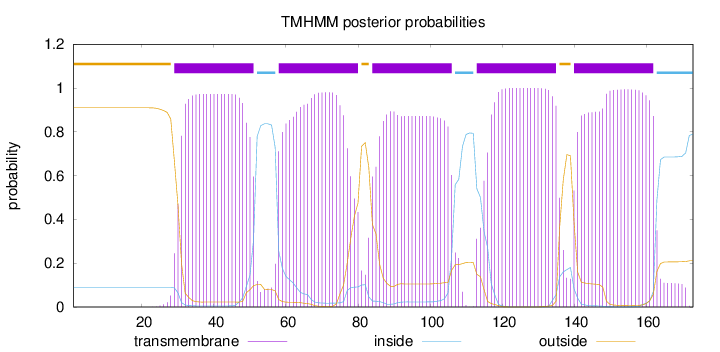
Topology
Length:
173
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
104.76035
Exp number, first 60 AAs:
23.18476
Total prob of N-in:
0.09031
POSSIBLE N-term signal
sequence
outside
1 - 28
TMhelix
29 - 51
inside
52 - 57
TMhelix
58 - 80
outside
81 - 83
TMhelix
84 - 106
inside
107 - 112
TMhelix
113 - 135
outside
136 - 139
TMhelix
140 - 162
inside
163 - 173
Population Genetic Test Statistics
Pi
265.477262
Theta
191.544122
Tajima's D
1.545961
CLR
0
CSRT
0.79886005699715
Interpretation
Uncertain
