Gene
KWMTBOMO15595
Pre Gene Modal
BGIBMGA010722
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.845
Sequence
CDS
ATGATAGCCAGAGAATACGAGGAGGAACCCAAAATTAGCGACTTGTGGGCCAGCAGAGGAAGCGTTAAAGCATTGGGTATATGCGTGTTCCTGGCGATGTTGCTGCAGTTATCCGGGATCGACGTGCTGTTATTCTACATGGAGGAGCTACTGGCGAAGGTCGGCACTAAGATCTCTCCGTCCGACGGAACTATCATCATGGGAGTCGTTCAGGTGGCGACGAGCTGTGTGACTCCGATGGTCGTGGATCGGCTGGGTCGGAAACTTCTCATGTGGACTACTTCGCTTGGACTTACTATATTTCTGACCTTAATCGGCGTTTACGCCCTTCTGGACAATTACTATAAATATGACGTCACATCCGTGACCTTCCTTCCGCTGCTGTGTCTAATTGTTTATATGATATTGTTCACTTTAGGTAAGTAA
Protein
MIAREYEEEPKISDLWASRGSVKALGICVFLAMLLQLSGIDVLLFYMEELLAKVGTKISPSDGTIIMGVVQVATSCVTPMVVDRLGRKLLMWTTSLGLTIFLTLIGVYALLDNYYKYDVTSVTFLPLLCLIVYMILFTLGK
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
A0A2H1VN09
A0A2A4JTW5
A0A194RQD1
A0A3S2LU80
A0A212EIU1
A0A0N0PA76
+ More
W0FW00 D7EK13 A0A1L8DIL5 A0A139WCV1 A0A1L8DIK8 A0A154PHJ7 D7EK14 A0A1L8DIL8 A0A1L8DIJ3 A8CWB1 A0A088ABY6 A0A0N0BDQ1 A0A182VIR9 A0A1Q3FQG8 A0A1Q3FQD3 A0A1Q3FQA6 A0A182TSI6 A0A1Y9J057 A0A182KX77 A0NBZ0 A0A1Q3FQH2 A0A182MM04 K7IP31 T1DG93 T1DE67 U5ESS0 A0A182QT48 A0A2A3E9H1 B0WZW7 A0A1Y9H2R2 A0A182S579 A0A182HJZ0 W5J543 A0A1Y0AWN8 Q16SU5 A0A232F7N7 A0A2M4A1M5 E0W0A9 A0A2M4A0H9 A0A2M4A0I2 A0A2M3ZF95 Q6NNE6 A0A182P8W7 A0A084VDW1 A0A194PK39 A0A182VS58 C1C3I4 A0A182T9M2 N6TVH8 A0A182GL69 D6X031 A0A182GVZ1 V5FVN9 E2B123 A0A1B0D4P3 A0A2M3ZFA3 A0A2M3ZFG5 A0A0L7RBX0 A0A1W4XJX7 J3JUQ1 A0A2M4BMJ9 A0A1W4XKU0 A0A1W4VHJ7 B4PVJ3 A0A182FKZ4 B3NYI2 Q7JVN6 A0A3B0JIU9 H9JMI9 A0A1W4UWZ5 A0A3B0K5K3 A0A3B0JXL8 A0A0T6BH34 A0A1W4W842 A0A182YLM9 A0A182J8U1 W4VS24 Q297W3 B4QXL4 B4I487 W4VSK2 A0A182S580 A0A1W4WJK3 A0A336M0J9 A0A151WM18 A0A1Y1M432 U5EQL0 D7EK15 V5I8R9 A0A1W4W7Z0
W0FW00 D7EK13 A0A1L8DIL5 A0A139WCV1 A0A1L8DIK8 A0A154PHJ7 D7EK14 A0A1L8DIL8 A0A1L8DIJ3 A8CWB1 A0A088ABY6 A0A0N0BDQ1 A0A182VIR9 A0A1Q3FQG8 A0A1Q3FQD3 A0A1Q3FQA6 A0A182TSI6 A0A1Y9J057 A0A182KX77 A0NBZ0 A0A1Q3FQH2 A0A182MM04 K7IP31 T1DG93 T1DE67 U5ESS0 A0A182QT48 A0A2A3E9H1 B0WZW7 A0A1Y9H2R2 A0A182S579 A0A182HJZ0 W5J543 A0A1Y0AWN8 Q16SU5 A0A232F7N7 A0A2M4A1M5 E0W0A9 A0A2M4A0H9 A0A2M4A0I2 A0A2M3ZF95 Q6NNE6 A0A182P8W7 A0A084VDW1 A0A194PK39 A0A182VS58 C1C3I4 A0A182T9M2 N6TVH8 A0A182GL69 D6X031 A0A182GVZ1 V5FVN9 E2B123 A0A1B0D4P3 A0A2M3ZFA3 A0A2M3ZFG5 A0A0L7RBX0 A0A1W4XJX7 J3JUQ1 A0A2M4BMJ9 A0A1W4XKU0 A0A1W4VHJ7 B4PVJ3 A0A182FKZ4 B3NYI2 Q7JVN6 A0A3B0JIU9 H9JMI9 A0A1W4UWZ5 A0A3B0K5K3 A0A3B0JXL8 A0A0T6BH34 A0A1W4W842 A0A182YLM9 A0A182J8U1 W4VS24 Q297W3 B4QXL4 B4I487 W4VSK2 A0A182S580 A0A1W4WJK3 A0A336M0J9 A0A151WM18 A0A1Y1M432 U5EQL0 D7EK15 V5I8R9 A0A1W4W7Z0
Pubmed
26354079
22118469
24391959
18362917
19820115
18194529
+ More
20966253 12364791 14747013 17210077 20075255 24330624 20920257 23761445 28341416 17510324 28648823 20566863 24438588 23537049 26483478 20798317 22516182 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 19121390 25244985 15632085 23185243 28004739
20966253 12364791 14747013 17210077 20075255 24330624 20920257 23761445 28341416 17510324 28648823 20566863 24438588 23537049 26483478 20798317 22516182 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 19121390 25244985 15632085 23185243 28004739
EMBL
ODYU01003431
SOQ42187.1
NWSH01000595
PCG75451.1
KQ459875
KPJ19609.1
+ More
RSAL01000221 RVE44063.1 AGBW02014588 OWR41380.1 KQ459078 KPJ03826.1 KF803265 AHF27419.1 DS497725 EFA12963.1 GFDF01007792 JAV06292.1 KQ971364 KYB25661.1 GFDF01007786 JAV06298.1 KQ434899 KZC10964.1 EFA12962.1 GFDF01007791 JAV06293.1 GFDF01007785 JAV06299.1 EU124608 ABV60326.1 KQ435851 KOX70761.1 GFDL01005309 JAV29736.1 GFDL01005268 JAV29777.1 GFDL01005255 JAV29790.1 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 GFDL01005283 JAV29762.1 AXCM01000233 GAMD01002846 JAA98744.1 GALA01001167 JAA93685.1 GANO01003127 JAB56744.1 AXCN02000090 KZ288338 PBC27839.1 DS232219 EDS37792.1 APCN01002224 ADMH02002130 ETN58528.1 KY921824 ART29413.1 CH477665 EAT37543.1 NNAY01000793 OXU26469.1 GGFK01001330 MBW34651.1 DS235858 EEB19065.1 GGFK01000993 MBW34314.1 GGFK01000934 MBW34255.1 GGFM01006410 MBW27161.1 BT011343 AAR96135.1 ATLV01011809 KE524723 KFB36155.1 KQ459601 KPI93687.1 BT081413 ACO51544.1 APGK01058785 KB741292 KI209303 ENN70297.1 ERL96014.1 JXUM01071368 JXUM01071369 KQ562660 KXJ75378.1 EFA10056.1 JXUM01091981 JXUM01091982 KQ563956 KXJ73021.1 GALX01006756 JAB61710.1 GL444771 EFN60596.1 AJVK01011560 GGFM01006414 MBW27165.1 GGFM01006409 MBW27160.1 KQ414617 KOC68300.1 BT126966 AEE61928.1 GGFJ01004857 MBW53998.1 CM000160 EDW97802.1 KRK03978.1 KRK03979.1 CH954181 EDV47963.1 AE014297 AY118796 AAF51941.1 AAM50656.1 OUUW01000005 SPP80252.1 BABH01016696 BABH01016697 SPP80251.1 SPP80250.1 LJIG01000288 KRT86652.1 GAPU01000003 JAB84614.1 CM000070 EAL28092.1 KRT00434.1 CM000364 EDX11800.1 CH480821 EDW55030.1 GAPU01000004 JAB84613.1 UFQT01000389 SSX23856.1 KQ982944 KYQ48914.1 GEZM01041155 JAV80582.1 GANO01004191 JAB55680.1 EFA12961.1 GALX01004205 JAB64261.1
RSAL01000221 RVE44063.1 AGBW02014588 OWR41380.1 KQ459078 KPJ03826.1 KF803265 AHF27419.1 DS497725 EFA12963.1 GFDF01007792 JAV06292.1 KQ971364 KYB25661.1 GFDF01007786 JAV06298.1 KQ434899 KZC10964.1 EFA12962.1 GFDF01007791 JAV06293.1 GFDF01007785 JAV06299.1 EU124608 ABV60326.1 KQ435851 KOX70761.1 GFDL01005309 JAV29736.1 GFDL01005268 JAV29777.1 GFDL01005255 JAV29790.1 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 GFDL01005283 JAV29762.1 AXCM01000233 GAMD01002846 JAA98744.1 GALA01001167 JAA93685.1 GANO01003127 JAB56744.1 AXCN02000090 KZ288338 PBC27839.1 DS232219 EDS37792.1 APCN01002224 ADMH02002130 ETN58528.1 KY921824 ART29413.1 CH477665 EAT37543.1 NNAY01000793 OXU26469.1 GGFK01001330 MBW34651.1 DS235858 EEB19065.1 GGFK01000993 MBW34314.1 GGFK01000934 MBW34255.1 GGFM01006410 MBW27161.1 BT011343 AAR96135.1 ATLV01011809 KE524723 KFB36155.1 KQ459601 KPI93687.1 BT081413 ACO51544.1 APGK01058785 KB741292 KI209303 ENN70297.1 ERL96014.1 JXUM01071368 JXUM01071369 KQ562660 KXJ75378.1 EFA10056.1 JXUM01091981 JXUM01091982 KQ563956 KXJ73021.1 GALX01006756 JAB61710.1 GL444771 EFN60596.1 AJVK01011560 GGFM01006414 MBW27165.1 GGFM01006409 MBW27160.1 KQ414617 KOC68300.1 BT126966 AEE61928.1 GGFJ01004857 MBW53998.1 CM000160 EDW97802.1 KRK03978.1 KRK03979.1 CH954181 EDV47963.1 AE014297 AY118796 AAF51941.1 AAM50656.1 OUUW01000005 SPP80252.1 BABH01016696 BABH01016697 SPP80251.1 SPP80250.1 LJIG01000288 KRT86652.1 GAPU01000003 JAB84614.1 CM000070 EAL28092.1 KRT00434.1 CM000364 EDX11800.1 CH480821 EDW55030.1 GAPU01000004 JAB84613.1 UFQT01000389 SSX23856.1 KQ982944 KYQ48914.1 GEZM01041155 JAV80582.1 GANO01004191 JAB55680.1 EFA12961.1 GALX01004205 JAB64261.1
Proteomes
UP000218220
UP000053240
UP000283053
UP000007151
UP000053268
UP000007266
+ More
UP000076502 UP000005203 UP000053105 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075883 UP000002358 UP000075886 UP000242457 UP000002320 UP000075884 UP000075900 UP000075840 UP000000673 UP000008820 UP000215335 UP000009046 UP000075885 UP000030765 UP000075920 UP000075901 UP000019118 UP000030742 UP000069940 UP000249989 UP000000311 UP000092462 UP000053825 UP000192223 UP000192221 UP000002282 UP000069272 UP000008711 UP000000803 UP000268350 UP000005204 UP000076408 UP000075880 UP000001819 UP000000304 UP000001292 UP000075809
UP000076502 UP000005203 UP000053105 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075883 UP000002358 UP000075886 UP000242457 UP000002320 UP000075884 UP000075900 UP000075840 UP000000673 UP000008820 UP000215335 UP000009046 UP000075885 UP000030765 UP000075920 UP000075901 UP000019118 UP000030742 UP000069940 UP000249989 UP000000311 UP000092462 UP000053825 UP000192223 UP000192221 UP000002282 UP000069272 UP000008711 UP000000803 UP000268350 UP000005204 UP000076408 UP000075880 UP000001819 UP000000304 UP000001292 UP000075809
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1VN09
A0A2A4JTW5
A0A194RQD1
A0A3S2LU80
A0A212EIU1
A0A0N0PA76
+ More
W0FW00 D7EK13 A0A1L8DIL5 A0A139WCV1 A0A1L8DIK8 A0A154PHJ7 D7EK14 A0A1L8DIL8 A0A1L8DIJ3 A8CWB1 A0A088ABY6 A0A0N0BDQ1 A0A182VIR9 A0A1Q3FQG8 A0A1Q3FQD3 A0A1Q3FQA6 A0A182TSI6 A0A1Y9J057 A0A182KX77 A0NBZ0 A0A1Q3FQH2 A0A182MM04 K7IP31 T1DG93 T1DE67 U5ESS0 A0A182QT48 A0A2A3E9H1 B0WZW7 A0A1Y9H2R2 A0A182S579 A0A182HJZ0 W5J543 A0A1Y0AWN8 Q16SU5 A0A232F7N7 A0A2M4A1M5 E0W0A9 A0A2M4A0H9 A0A2M4A0I2 A0A2M3ZF95 Q6NNE6 A0A182P8W7 A0A084VDW1 A0A194PK39 A0A182VS58 C1C3I4 A0A182T9M2 N6TVH8 A0A182GL69 D6X031 A0A182GVZ1 V5FVN9 E2B123 A0A1B0D4P3 A0A2M3ZFA3 A0A2M3ZFG5 A0A0L7RBX0 A0A1W4XJX7 J3JUQ1 A0A2M4BMJ9 A0A1W4XKU0 A0A1W4VHJ7 B4PVJ3 A0A182FKZ4 B3NYI2 Q7JVN6 A0A3B0JIU9 H9JMI9 A0A1W4UWZ5 A0A3B0K5K3 A0A3B0JXL8 A0A0T6BH34 A0A1W4W842 A0A182YLM9 A0A182J8U1 W4VS24 Q297W3 B4QXL4 B4I487 W4VSK2 A0A182S580 A0A1W4WJK3 A0A336M0J9 A0A151WM18 A0A1Y1M432 U5EQL0 D7EK15 V5I8R9 A0A1W4W7Z0
W0FW00 D7EK13 A0A1L8DIL5 A0A139WCV1 A0A1L8DIK8 A0A154PHJ7 D7EK14 A0A1L8DIL8 A0A1L8DIJ3 A8CWB1 A0A088ABY6 A0A0N0BDQ1 A0A182VIR9 A0A1Q3FQG8 A0A1Q3FQD3 A0A1Q3FQA6 A0A182TSI6 A0A1Y9J057 A0A182KX77 A0NBZ0 A0A1Q3FQH2 A0A182MM04 K7IP31 T1DG93 T1DE67 U5ESS0 A0A182QT48 A0A2A3E9H1 B0WZW7 A0A1Y9H2R2 A0A182S579 A0A182HJZ0 W5J543 A0A1Y0AWN8 Q16SU5 A0A232F7N7 A0A2M4A1M5 E0W0A9 A0A2M4A0H9 A0A2M4A0I2 A0A2M3ZF95 Q6NNE6 A0A182P8W7 A0A084VDW1 A0A194PK39 A0A182VS58 C1C3I4 A0A182T9M2 N6TVH8 A0A182GL69 D6X031 A0A182GVZ1 V5FVN9 E2B123 A0A1B0D4P3 A0A2M3ZFA3 A0A2M3ZFG5 A0A0L7RBX0 A0A1W4XJX7 J3JUQ1 A0A2M4BMJ9 A0A1W4XKU0 A0A1W4VHJ7 B4PVJ3 A0A182FKZ4 B3NYI2 Q7JVN6 A0A3B0JIU9 H9JMI9 A0A1W4UWZ5 A0A3B0K5K3 A0A3B0JXL8 A0A0T6BH34 A0A1W4W842 A0A182YLM9 A0A182J8U1 W4VS24 Q297W3 B4QXL4 B4I487 W4VSK2 A0A182S580 A0A1W4WJK3 A0A336M0J9 A0A151WM18 A0A1Y1M432 U5EQL0 D7EK15 V5I8R9 A0A1W4W7Z0
PDB
6N3I
E-value=3.274e-05,
Score=106
Ontologies
GO
Topology
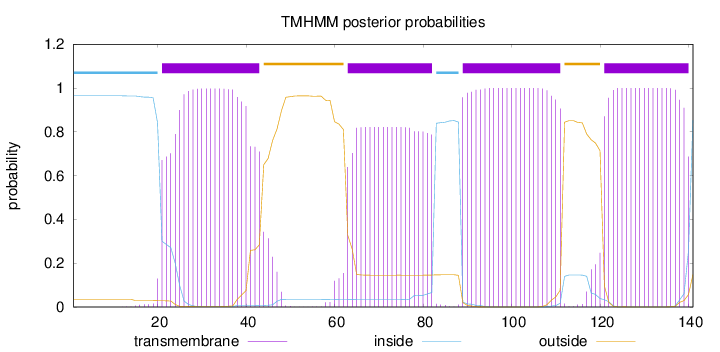
Length:
141
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
81.26118
Exp number, first 60 AAs:
22.14289
Total prob of N-in:
0.96511
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 62
TMhelix
63 - 82
inside
83 - 88
TMhelix
89 - 111
outside
112 - 120
TMhelix
121 - 140
inside
141 - 141
Population Genetic Test Statistics
Pi
260.721847
Theta
180.509164
Tajima's D
1.169803
CLR
0.138462
CSRT
0.705214739263037
Interpretation
Uncertain
